Direct reprogramming of stem cell properties in colon cancer cells by CD44 - PubMed (original) (raw)
Direct reprogramming of stem cell properties in colon cancer cells by CD44
Ying-Jhen Su et al. EMBO J. 2011.
Abstract
Cancer progression is commonly segregated into processes of primary tumour growth and secondary metastasis. Recent evidence suggests that a subpopulation of cancer cells, cancer stem cells (CSCs), is responsible for tumour growth in cancer. However, the role of CSCs in cancer metastasis is unclear. In this study, we found that the C terminus of CD44 contributes to sphere formation and survival in vitro via the CD44-SRC-integrin axis. In addition, nuclear CD44/acetylated-STAT3 is required for clonal formation in vitro and tumourigenicity in vivo. Nuclear CD44 binds to various promoters identified by chromatin immunoprecipitation-seq, including that of c-myc and Twist1, leading to cell fate change through transcriptional reprogramming. We propose that nuclear CD44/acetylated-STAT3 performs an unexpected tumour-progressing function by enhancing cell outgrowth into structures where cells with properties of CSCs can be generated from differentiated somatic cells in suspension culture, and then exhibit attributes of cells that have undergone an epithelial-mesenchymal transition, leading to tumour metastasis, and a resulting worse prognosis.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
Figure 1
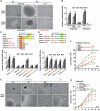
CD44 allows outgrowth of cells into spheres, leading to stable changes in cell proliferative ability and morphology after the suspension culture. (A) Microscopic analysis of spheres cultivated in suspension for 6 and 12 days. (Top panel) HT29/CD44− cells were transfected with plasmid encoding CD44s (CD44-myc) or control vector (Mock). (Bottom panel) HT29/CD44+ cells were infected by lentivirus-encoding shRNA targeting CD44 (CD44-shRNA) or control scrambled shRNA (Cont-shRNA). Bars, 50 μm. (B) In vitro quantification of spheres formed by cells described in (A) during four serial passages (p1–p4). (C, D) The C terminus of CD44 is required for sphere formation over four serial passages. (Top) Schematic representation of the transcripts encoding wild-type CD44s and its in-frame C-terminal (in C)/N-terminal (in D) deletion mutants. TM, transmembrane domain; ICD, intracellular domain. (Bottom) In vitro quantification of spheres formed by stable HT29/CD44− clones expressing C-terminal deletion mutants of CD44s. (E) The sphere-forming culture triggers stable changes in cell morphology. Higher-power views are shown in the bottom panels. Bars, 50 μm. (F, G) The sphere-forming culture triggers stable changes in cell ability. HT29/CD44− stable clones (in F) and HT29/CD44+ cells (in G) (described in E) were plated at 105 cells per six-well dish in 1% FBS RPMI medium. Total viable cell number was determined. Data in (B–D, F, G) were derived from three independent experiments and are presented as mean values±s.d. *P<0.05; **P<0.01; ***P<0.005 (_t_-test). AD (in E–G): monolayer cell culture grown in tissue culture plates; SPH (in E–G): cultured under sphere-forming conditions; SPH → AD (in E–G): all cells in the spheres migrated back onto the plate to reform a monolayer.
Figure 2
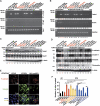
C terminus of CD44 leads to increased resistance to anoikis during the sphere-forming culture through the CD44–SRC–integrin axis in lipid rafts. (A) Cells after treatment were then cultured in suspension for 120 h before apoptosis assays by flow cytometric analyses of sub-G1 fractions. (B, C) Stable HT29/CD44− clones expressing C-terminal deletion mutants of CD44s (in B) or N-terminal deletion mutants of CD44s (in C) were cultured in suspension for 120 h before apoptosis assays by flow cytometric analyses of sub-G1 fractions. (D, E) HT29/CD44+ (in D) and HT29/CD44−/CD44-myc (in E) cells were cultured (monolayer) in tissue culture plates (AD, left panel) or in sphere-forming culture (SPH, right panel). Triton X-100-insoluble raft fractions were isolated by sucrose gradient fractionation, pooled, and analysed by western blotting for the individual proteins indicated. (F) Stable clones were cultured (monolayer) in tissue culture plates (AD, left panel) or in sphere-forming culture (SPH, right panel). The lipid rafts were isolated by sucrose gradient fractionation. An equal volume of each fraction was analysed by western blotting for the proteins indicated. The relative intensities of the bands were densitometrically quantified and normalized to Cont-shRNA (CD44+ cells) and Mock (CD44− cells). The red asterisks in upper part: CD44 proteins with expected sizes. (G) HT29/CD44+ cells were treated with control IgG or blocking Ab against integrin β1 or infected with a lentivirus encoding a shRNA targeting Src (Src-shRNA) or a control scrambled shRNA (Cont-shRNA), followed by cultured in suspension for 120 h, and apoptosis was measured by flow cytometric analysis of sub-G1 fractions. (H) In vitro quantification of spheres formed by cells described in (G) during four serial passages (p4). Data in (A–C, G, H) were derived from three independent experiments and are presented as mean values±s.d. *P<0.05; **P<0.01; ***P<0.005 (_t_-test).
Figure 3
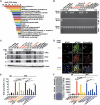
Nuclear CD44/acetylated-STAT3 is required for sphere growth in vitro in sphere-forming cells. (A) HT29/CD44− (expressing CD44s mutants) stable clones were cultured in sphere-forming conditions for 12 days, immunostained for CD44 (green, top panel) and counterstained with DAPI (blue, second panel); immunostained for STAT3 (red, third panel) and counterstained with DAPI (blue, bottom panel). Representative images taken by confocal laser microscopy are shown. Bars, 50 μm. (B) Nuclear extracts were prepared from stable HT29/CD44− clones expressing wild-type CD44s or the NLS mutant that were cultured in suspension for the indicated times. 12D/SPH: cells were cultured in sphere-forming conditions for 12 days; 120/Ran(Q69L): cells were transfected with plasmids encoding Ran(Q69L) and then cultured in suspension for 120 h; TCL, total cell lysate. (C) (Top panel) Nuclear extracts were prepared from spheres of HT29 stable clones and then immunoprecipitated using anti-CD44 followed by western blotting. (Bottom panel) The diameter of spheres was measured after 12 days in sphere-forming culture. The red asterisks in upper part: CD44 proteins with expected sizes. ND, not determined. (D, E) Stable HT29/CD44− clones infected with a lentivirus encoding a shRNA targeting STAT3 or a control scrambled shRNA (in D) and expressing wild-type (in D, E) or mutant (in E) CD44s were cultured in suspension for the designated times before apoptosis assays by flow cytometric analyses of sub-G1 fractions. (F, G) The cells described in (D, E) were cultured in sphere-forming conditions for the indicated times; the total viable cell number was determined. (H) Stable clones were cultured in suspension with the indicated concentrations of serum for 12 days; the doubling time was determined. (I) Stable clones were cultured in sphere-forming conditions for 12 days. The number of spheres/100 cells and the size of spheres were determined. Data in (C–I) were derived from three independent experiments and are presented as mean values±s.d. *P<0.05; **P<0.01; ***P<0.005 (_t_-test). WT/AD (in C): HT29/CD44− cells expressing wild-type CD44s were cultured (monolayer) in tissue culture plates. SPH (in B, C): cultured in sphere-forming conditions.
Figure 4
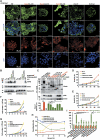
CD44-expressing cells are reprogrammed into stem-like cells after the suspension culture. (A) Validation by qChIP-PCR of putative nuclear CD44 target genes in HT29/CD44+ spheres. A ChIP assay was performed with chromatin from HT29/CD44+ spheres using anti-CD44 mAb. The immunoprecipitated DNA was amplified by qPCR. Equal amounts of anti-CD44 ChIP DNA and total input DNA were used for qPCR employing SYBR Green detection with an ABI7900HT system. (B) Nuclear extracts were prepared from spheres of HT29/CD44+ infected with a lentivirus encoding a shRNA targeting CD44 or a control scrambled shRNA and those of the HT29/CD44−/CD44-myc mutants infected with a lentivirus encoding a shRNA targeting STAT3 or p300, a control scrambled shRNA, or expressing HDAC1. ChIP was performed using anti-CD44 mAb or control IgG. PCR amplification of the designated regions within the c-myc promoter was performed. (C) Total cell lysates were prepared from the spheres described in (B). The expression of c-Myc, SOX2, and OCT4 was assessed by western blotting. β-Actin was used as a loading control. (D) Immunostaining for SOX2 and OCT4 is shown in spheres of HT29/CD44− expressing wild-type CD44s or the NLS mut. (E) Cells derived from spheres described in Figure 3I were stained with Hoechst 33342. Side-population cells excluding Hoechst 33342 were determined. (F) Anchorage-independent assays (AIGs) were performed on cells derived from spheres described in Figure 3I. Data in (A, E, F) were derived from three independent experiments and are presented as mean values±s.d. **P<0.01; ***P<0.005 (_t_-test). WT/AD (in B–D and F): HT29/CD44− cells expressing wild-type CD44s were cultured (monolayer) in tissue culture plates. AD (in E, F): monolayer culture in tissue culture plates; SPH (in D–F): cultured under sphere-forming conditions.
Figure 5
CD44-expressing cells after the suspension culture exhibit attributes of cells that have undergone an EMT. (A) Nuclear extracts were prepared from spheres described in Figure 4B. ChIP was performed using anti-CD44 or control IgG. PCR amplification of the designated regions within the Twist1 promoter was performed. (B) mRNAs were prepared from the spheres described in Figure 4B. The expression of Twist1 and Snail1 was evaluated by RT–PCR. (C) Total cell lysates were prepared from the spheres described in Figure 4B. The expression of Twist1 and Snail1 was measured by western blotting. (D) Total cell lysates were prepared from the spheres described in Figure 4B. The expression of epithelial markers (P-cadherin and claudin-1) and mesenchymal markers (Vimentin and Fibronectin) was measured by western blotting. (E) Immunostaining for P-cadherin and Vimentin was shown in spheres of HT29/CD44− expressing wild-type CD44s or the NLS mut. (F) Boyden chamber assays were performed on cells derived from spheres described in Figure 2I to assess the invasive activity. Data were derived from three independent experiments and are presented as mean values±s.d. *P<0.05; **P<0.01 (_t_-test). The relative intensities of the bands (in B–D) were densitometrically quantified and normalized to Cont-shRNA (CD44+ cells) and WT/AD (CD44− cells). WT/AD (in A–E): HT29/CD44− cells expressing wild-type CD44s were cultured (monolayer) in tissue culture plates. AD (in F): monolayer culture in tissue culture plates; SPH (in F): cultured under sphere-forming conditions.
Figure 6
Characterization of the highly metastatic cancer cells derived from the xenografts. (A, B) Real-time PCR showing expression of mRNAs for the CSCs-related genes (in A) and EMT-related genes (in B) in the highly metastatic cancer cells derived from the xenografts (HT29/CD44+/Cont-shRNA). The comparison is to cells in subconfluent monolayers. (C) Cells derived from the xenografts described in (A) were stained with Hoechst 33342. Side-population cells excluding Hoechst 33342 were determined. (D) Anchorage-independent assays (AIGs) were performed on cells derived from the xenografts described in (A). (E) Model proposing a pathway for generation of cells with properties of CSCs from differentiated somatic cells in the suspension condition, and then exhibit attributes of cells that have undergone an EMT, leading to tumour metastasis. See text for discussion. Data in (A–D) were derived from three independent experiments and are presented as mean values±s.d.
Similar articles
- CD44 and CD24 coordinate the reprogramming of nasopharyngeal carcinoma cells towards a cancer stem cell phenotype through STAT3 activation.
Shen YA, Wang CY, Chuang HY, Hwang JJ, Chi WH, Shu CH, Ho CY, Li WY, Chen YJ. Shen YA, et al. Oncotarget. 2016 Sep 6;7(36):58351-58366. doi: 10.18632/oncotarget.11113. Oncotarget. 2016. PMID: 27521216 Free PMC article. - Maintenance of the stemness in CD44(+) HCT-15 and HCT-116 human colon cancer cells requires miR-203 suppression.
Ju SY, Chiou SH, Su Y. Ju SY, et al. Stem Cell Res. 2014 Jan;12(1):86-100. doi: 10.1016/j.scr.2013.09.011. Epub 2013 Oct 2. Stem Cell Res. 2014. PMID: 24145190 - Activation of CD44-Lipoprotein lipase axis in breast cancer stem cells promotes tumorigenesis.
Manupati K, Yeeravalli R, Kaushik K, Singh D, Mehra B, Gangane N, Gupta A, Goswami K, Das A. Manupati K, et al. Biochim Biophys Acta Mol Basis Dis. 2021 Nov 1;1867(11):166228. doi: 10.1016/j.bbadis.2021.166228. Epub 2021 Jul 24. Biochim Biophys Acta Mol Basis Dis. 2021. PMID: 34311079 - Activation of Matrix Hyaluronan-Mediated CD44 Signaling, Epigenetic Regulation and Chemoresistance in Head and Neck Cancer Stem Cells.
Bourguignon LYW, Earle C, Shiina M. Bourguignon LYW, et al. Int J Mol Sci. 2017 Aug 24;18(9):1849. doi: 10.3390/ijms18091849. Int J Mol Sci. 2017. PMID: 28837080 Free PMC article. Review. - Hyaluronan-CD44 axis orchestrates cancer stem cell functions.
Skandalis SS, Karalis TT, Chatzopoulos A, Karamanos NK. Skandalis SS, et al. Cell Signal. 2019 Nov;63:109377. doi: 10.1016/j.cellsig.2019.109377. Epub 2019 Jul 27. Cell Signal. 2019. PMID: 31362044 Review.
Cited by
- Understanding cancer stem cell heterogeneity and plasticity.
Tang DG. Tang DG. Cell Res. 2012 Mar;22(3):457-72. doi: 10.1038/cr.2012.13. Epub 2012 Jan 17. Cell Res. 2012. PMID: 22357481 Free PMC article. Review. - The role of lipid rafts in cancer cell adhesion and migration.
Murai T. Murai T. Int J Cell Biol. 2012;2012:763283. doi: 10.1155/2012/763283. Epub 2011 Dec 29. Int J Cell Biol. 2012. PMID: 22253629 Free PMC article. - Cellular Effects of Selected Unsymmetrical Bisacridines on the Multicellular Tumor Spheroids of HCT116 Colon and A549 Lung Cancer Cells in Comparison to Monolayer Cultures.
Kulesza J, Paluszkiewicz E, Augustin E. Kulesza J, et al. Int J Mol Sci. 2023 Oct 30;24(21):15780. doi: 10.3390/ijms242115780. Int J Mol Sci. 2023. PMID: 37958764 Free PMC article. - An aberrant nuclear localization of E-cadherin is a potent inhibitor of Wnt/β-catenin-elicited promotion of the cancer stem cell phenotype.
Su YJ, Chang YW, Lin WH, Liang CL, Lee JL. Su YJ, et al. Oncogenesis. 2015 Jun 15;4(6):e157. doi: 10.1038/oncsis.2015.17. Oncogenesis. 2015. PMID: 26075748 Free PMC article. - CD44 splice isoform switching determines breast cancer stem cell state.
Zhang H, Brown RL, Wei Y, Zhao P, Liu S, Liu X, Deng Y, Hu X, Zhang J, Gao XD, Kang Y, Mercurio AM, Goel HL, Cheng C. Zhang H, et al. Genes Dev. 2019 Feb 1;33(3-4):166-179. doi: 10.1101/gad.319889.118. Epub 2019 Jan 28. Genes Dev. 2019. PMID: 30692202 Free PMC article.
References
- Ansieau S, Bastid J, Doreau A, Morel AP, Bouchet BP, Thomas C, Fauvet F, Puisieux I, Doglioni C, Piccinin S, Maestro R, Voeltzel T, Selmi A, Valsesia-Wittmann S, Caron de Fromentel C, Puisieux A (2008) Induction of EMT by twist proteins as a collateral effect of tumor-promoting inactivation of premature senescence. Cancer Cell 14: 79–89 - PubMed
- Challen GA, Little MH (2006) A side order of stem cells: the SP phenotype. Stem Cells 24: 3–12 - PubMed
- Chambers AF, Groom AC, MacDonald IC (2002) Dissemination and growth of cancer cells in metastatic sites. Nat Rev Cancer 2: 563–572 - PubMed
- Collins AT, Berry PA, Hyde C, Stower MJ, Maitland NJ (2005) Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res 65: 10946–10951 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous