Atomic structure of bacteriophage Sf6 tail needle knob - PubMed (original) (raw)
Atomic structure of bacteriophage Sf6 tail needle knob
Anshul Bhardwaj et al. J Biol Chem. 2011.
Abstract
Podoviridae are double-stranded DNA bacteriophages that use short, non-contractile tails to adsorb to the host cell surface. Within the tail apparatus of P22-like phages, a dedicated fiber known as the "tail needle" likely functions as a cell envelope-penetrating device to promote ejection of viral DNA inside the host. In Sf6, a P22-like phage that infects Shigella flexneri, the tail needle presents a C-terminal globular knob. This knob, absent in phage P22 but shared in other members of the P22-like genus, represents the outermost exposed tip of the virion that contacts the host cell surface. Here, we report a crystal structure of the Sf6 tail needle knob determined at 1.0 Å resolution. The structure reveals a trimeric globular domain of the TNF fold structurally superimposable with that of the tail-less phage PRD1 spike protein P5 and the adenovirus knob, domains that in both viruses function in receptor binding. However, P22-like phages are not known to utilize a protein receptor and are thought to directly penetrate the host surface. At 1.0 Å resolution, we identified three equivalents of l-glutamic acid (l-Glu) bound to each subunit interface. Although intimately bound to the protein, l-Glu does not increase the structural stability of the trimer nor it affects its ability to self-trimerize in vitro. In analogy to P22 gp26, we suggest the tail needle of phage Sf6 is ejected through the bacterial cell envelope during infection and its C-terminal knob is threaded through peptidoglycan pores formed by glycan strands.
Figures
FIGURE 1.
Bacteriophage Sf6 tail needle forms an elongated fiber with a C-terminal globular domain. A, micrographs of negatively stained Sf6 tail needle fibers. B, five class averages resulting from several iterations of multireference alignment (24). The approximate length of the Sf6 tail needle estimated from the projection average is ∼220 Å. C, schematic diagram of the Sf6 tail needle (residues 1–282) and the knob construct used in this study (residues 132–282).
FIGURE 2.
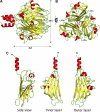
Atomic structure of bacteriophage Sf6 tail needle knob bound to l-Glu visualized crystallographically at 1.0 Å resolution. A and B, ribbon diagram of the trimeric Sf6 tail needle knob shown in side and top views, respectively. The structure is colored by secondary structure elements with β-strands and α-helices in yellow and red, respectively.
l
-Glu and phosphate ion are shown as ball-and-stick diagrams. C, ribbon diagram of the tail needle knob monomer reveals the characteristic architecture of a TNF jellyroll fold consisting of eight β-strands organized in two β-sheets with a tilt angle of 36°.
FIGURE 3.
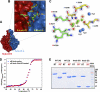
l-Glu binding cleft in Sf6 tail needle knob. A, surface representation of Sf6 knob highlighting the interface between subunits A and B (colored in blue and red, respectively). B, zoom-in view of the subunit interface where
l
-Glu is bound. Subunit A and B side chains from the final refined model are shown in sticks, and
l
-Glu is shown in a stick-and-ball representation. C, the final refined model for
l
-Glu and the interacting residues from both subunits are overlaid to the final 2_Fo_ − Fc electron density map calculated at 1.0 Å resolution and contoured at 1.5σ above background (in blue). The
l
-Glu is bound in a cleft formed at the interface of two knob monomers, closely surrounded by B-Glu146, B-Lys277, A-Leu255, B-Lys200, A-Ser248, and A-Asp250. D, thermal stability of the native and GdnHCl-treated Sf6 knob. Both samples unfold irreversibly with a melting temperature (Tm) of ∼67 °C. E, SDS resistance assay of wild type Sf6 tail needle and Sf6 knob that were previously treated under native conditions (N) or dialyzed against 1.5
m
GdnHCl, 0.5
m
NaCl (D). RT and 95° denote samples that were either incubated at 22 °C or boiled for 5 min prior to being electrophoresed.
FIGURE 4.
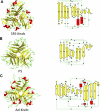
Comparison of the Sf6 tail needle knob with the receptor-binding domains of PRD1 and adenovirus. Left panel, Sf6 knob (A); PRD1 P5 (PDB code 1YQ5) (B); and adenovirus knob (PDB code 1NOB) (C). Right panel, topological diagrams of each protomer (generated using PDBsum (50)). Red and yellow are α-helices and β-strands, respectively.
FIGURE 5.
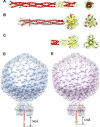
Modeling bacteriophage tail needles. A, crystal structure of P22 tail needle gp26 (PDB code 3C9I). B, homology model of the full-length Sf6 tail needle. The portion of the tail needle helical core missing (residues 1–132) in the construct used for crystallography was modeled using Phyre (38) and is highlighted in gray. C, crystal structure of the receptor binding protein from lactococcal phage TP901-1 base plate (PDB code 3EJC) (20, 39). D, a composite model of bacteriophage P22 and Sf6 (E) mature virions generated by fitting the atomic structure of gp26 tail needles and tailspikes into the asymmetric cryo-EM reconstruction of mature P22 (EMDB ID 1220) (6).
FIGURE 6.
A structural model for Sf6 tail needle penetration of Shigella cell wall. Top (A), tilted (B), and side (C) view of the Sf6 tail needle embedded into a pore of the Gram-negative cell wall, according to the structure of PG determined by Meroueh et al. (49). Glycan strands are colored orange, and the cross-linking stem peptides are colored green, with
d
-Glu in purple. The Sf6 tail needle is shown as a transparent solvent-accessible gray surface overlaid to the ribbon structure.
Similar articles
- The tip of the tail needle affects the rate of DNA delivery by bacteriophage P22.
Leavitt JC, Gogokhia L, Gilcrease EB, Bhardwaj A, Cingolani G, Casjens SR. Leavitt JC, et al. PLoS One. 2013 Aug 12;8(8):e70936. doi: 10.1371/journal.pone.0070936. eCollection 2013. PLoS One. 2013. PMID: 23951045 Free PMC article. - Structural Plasticity of the Protein Plug That Traps Newly Packaged Genomes in Podoviridae Virions.
Bhardwaj A, Sankhala RS, Olia AS, Brooke D, Casjens SR, Taylor DJ, Prevelige PE Jr, Cingolani G. Bhardwaj A, et al. J Biol Chem. 2016 Jan 1;291(1):215-26. doi: 10.1074/jbc.M115.696260. Epub 2015 Nov 16. J Biol Chem. 2016. PMID: 26574546 Free PMC article. - An evolutionarily conserved family of virion tail needles related to bacteriophage P22 gp26: correlation between structural stability and length of the alpha-helical trimeric coiled coil.
Bhardwaj A, Walker-Kopp N, Casjens SR, Cingolani G. Bhardwaj A, et al. J Mol Biol. 2009 Aug 7;391(1):227-45. doi: 10.1016/j.jmb.2009.05.069. Epub 2009 May 29. J Mol Biol. 2009. PMID: 19482036 Free PMC article. - Structures of the tailed bacteriophages that infect Gram-positive bacteria.
Huang L, Xiang Y. Huang L, et al. Curr Opin Virol. 2020 Dec;45:65-74. doi: 10.1016/j.coviro.2020.09.002. Epub 2020 Nov 1. Curr Opin Virol. 2020. PMID: 33142120 Review. - Short noncontractile tail machines: adsorption and DNA delivery by podoviruses.
Casjens SR, Molineux IJ. Casjens SR, et al. Adv Exp Med Biol. 2012;726:143-79. doi: 10.1007/978-1-4614-0980-9_7. Adv Exp Med Biol. 2012. PMID: 22297513 Review.
Cited by
- High-resolution cryo-EM structure of the Shigella virus Sf6 genome delivery tail machine.
Li F, Hou CD, Yang R, Whitehead R 3rd, Teschke CM, Cingolani G. Li F, et al. Sci Adv. 2022 Dec 9;8(49):eadc9641. doi: 10.1126/sciadv.adc9641. Epub 2022 Dec 7. Sci Adv. 2022. PMID: 36475795 Free PMC article. - Ecology, Structure, and Evolution of Shigella Phages.
Subramanian S, Parent KN, Doore SM. Subramanian S, et al. Annu Rev Virol. 2020 Sep 29;7(1):121-141. doi: 10.1146/annurev-virology-010320-052547. Epub 2020 May 11. Annu Rev Virol. 2020. PMID: 32392456 Free PMC article. Review. - Use of Localized Reconstruction to Visualize the Shigella Phage Sf6 Tail Apparatus.
Hou CD, Li F, Iglesias S, Cingolani G. Hou CD, et al. Methods Mol Biol. 2024;2738:215-228. doi: 10.1007/978-1-0716-3549-0_14. Methods Mol Biol. 2024. PMID: 37966602 Free PMC article. - OmpA and OmpC are critical host factors for bacteriophage Sf6 entry in Shigella.
Parent KN, Erb ML, Cardone G, Nguyen K, Gilcrease EB, Porcek NB, Pogliano J, Baker TS, Casjens SR. Parent KN, et al. Mol Microbiol. 2014 Apr;92(1):47-60. doi: 10.1111/mmi.12536. Epub 2014 Mar 6. Mol Microbiol. 2014. PMID: 24673644 Free PMC article. - Cryo-EM structure of the periplasmic tunnel of T7 DNA-ejectosome at 2.7 Å resolution.
Swanson NA, Lokareddy RK, Li F, Hou CD, Leptihn S, Pavlenok M, Niederweis M, Pumroy RA, Moiseenkova-Bell VY, Cingolani G. Swanson NA, et al. Mol Cell. 2021 Aug 5;81(15):3145-3159.e7. doi: 10.1016/j.molcel.2021.06.001. Epub 2021 Jul 1. Mol Cell. 2021. PMID: 34214465 Free PMC article.
References
- Ackermann H. W. (2003) Res. Microbiol. 154, 245–251 - PubMed
- Casjens S. R., Thuman-Commike P. A. (2011) Virology 411, 393–415 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 1R56AI076509-01A1/AI/NIAID NIH HHS/United States
- R01 AI074825/AI/NIAID NIH HHS/United States
- R56 AI076509/AI/NIAID NIH HHS/United States
- R56 AI074825/AI/NIAID NIH HHS/United States
- AI074825/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Miscellaneous