A public genome-scale lentiviral expression library of human ORFs - PubMed (original) (raw)
. 2011 Jun 26;8(8):659-61.
doi: 10.1038/nmeth.1638.
Jesse S Boehm, Xinping Yang, Kourosh Salehi-Ashtiani, Tong Hao, Yun Shen, Rakela Lubonja, Sapana R Thomas, Ozan Alkan, Tashfeen Bhimdi, Thomas M Green, Cory M Johannessen, Serena J Silver, Cindy Nguyen, Ryan R Murray, Haley Hieronymus, Dawit Balcha, Changyu Fan, Chenwei Lin, Lila Ghamsari, Marc Vidal, William C Hahn, David E Hill, David E Root
Affiliations
- PMID: 21706014
- PMCID: PMC3234135
- DOI: 10.1038/nmeth.1638
A public genome-scale lentiviral expression library of human ORFs
Xiaoping Yang et al. Nat Methods. 2011.
Abstract
Functional characterization of the human genome requires tools for systematically modulating gene expression in both loss-of-function and gain-of-function experiments. We describe the production of a sequence-confirmed, clonal collection of over 16,100 human open-reading frames (ORFs) encoded in a versatile Gateway vector system. Using this ORFeome resource, we created a genome-scale expression collection in a lentiviral vector, thereby enabling both targeted experiments and high-throughput screens in diverse cell types.
Figures
Figure 1. Overview of hORFeome V8.1
(a) Schematic of hORFeome V8.1 creation. Templates from the Mammalian Gene Collection (MGC) were transferred into the Gateway system via PCR and recombinational cloning, resolved as clonal isolates, fully sequenced and rearrayed. (b) Sequencing outcomes for 19,281 ORF samples in polyclonal format, from which single colonies were isolated. 14,524 ORFs were fully sequenced and accepted into the final collection (Complete, accepted). 198 ORFs were fully sequenced, but rejected for lacking a start codon (Complete, rejected). 825 ORFs were partially sequenced, including undetermined nucleotides (Partial). 823 ORFs were made clonal but were intentionally not sequenced, since these ORFs were isoforms of other ORFs in the sequencing pool and could cause unambiguous read mapping (Not attempted). See Supplementary Figure 3 for more details. (c) Alignment of the 14,524 completely sequenced clones with MGC templates. 12,736 clones have identical sequence as templates or have one synonymous error only (Perfect), and another 1,788 clones have additional mutations (Mutant). (d) Alignment of the 14,524 completely sequenced clones with NCBI RefSeq transcripts. 10,216 ORFs represent full length coding sequences with > 99% homology (Full), 1,545 ORFs were partial length coding sequences with > 85% homology (Partial) and 2,763 clones fell into other categories (Other). See Supplementary Figure 4 for more details.
Figure 2. Overview and performance of the CCSB-Broad Lentiviral Expression Library
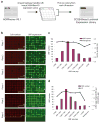
(a) Schematic of the creation of the CCSB-Broad Lentiviral Expression Library. pLX304-Blast-V5 is a custom lentiviral vector validated for high-throughput screening encoding Blasticidin (Blast) resistance and a C-terminal V5-epitope tag. (b) Evaluation of high-throughput ORF transduction and expression in A549 lung cancer cell lines. The micrographs show images of cells stained for cellular DNA (to assess cell number) and with antibodies recognizing the V5 epitope (to assess ORF expression) after lentiviral infection and three days of growth in blasticidin. Wells in which no virus was added are highlighted with yellow outline. (c) Distribution of ORF sizes and average viral titer as a function of ORF size. (d) ORF expression as a function of ORF size. ORFs larger than 3 kb showed a decreased yet detectable above-background level of expression. Background was assessed from cells expressing a control vector without V5 expression.
Similar articles
- Generation of a genome scale lentiviral vector library for EF1α promoter-driven expression of human ORFs and identification of human genes affecting viral titer.
Škalamera D, Dahmer M, Purdon AS, Wilson BM, Ranall MV, Blumenthal A, Gabrielli B, Gonda TJ. Škalamera D, et al. PLoS One. 2012;7(12):e51733. doi: 10.1371/journal.pone.0051733. Epub 2012 Dec 12. PLoS One. 2012. PMID: 23251614 Free PMC article. - The full-ORF clone resource of the German cDNA Consortium.
Bechtel S, Rosenfelder H, Duda A, Schmidt CP, Ernst U, Wellenreuther R, Mehrle A, Schuster C, Bahr A, Blöcker H, Heubner D, Hoerlein A, Michel G, Wedler H, Köhrer K, Ottenwälder B, Poustka A, Wiemann S, Schupp I. Bechtel S, et al. BMC Genomics. 2007 Oct 31;8:399. doi: 10.1186/1471-2164-8-399. BMC Genomics. 2007. PMID: 17974005 Free PMC article. - A high-throughput platform for lentiviral overexpression screening of the human ORFeome.
Škalamera D, Ranall MV, Wilson BM, Leo P, Purdon AS, Hyde C, Nourbakhsh E, Grimmond SM, Barry SC, Gabrielli B, Gonda TJ. Škalamera D, et al. PLoS One. 2011;6(5):e20057. doi: 10.1371/journal.pone.0020057. Epub 2011 May 24. PLoS One. 2011. PMID: 21629697 Free PMC article. - Genome-scale loss-of-function screening with a lentiviral RNAi library.
Root DE, Hacohen N, Hahn WC, Lander ES, Sabatini DM. Root DE, et al. Nat Methods. 2006 Sep;3(9):715-9. doi: 10.1038/nmeth924. Nat Methods. 2006. PMID: 16929317 Review. - New insights into chemical biology from ORFeome libraries.
Yashiroda Y, Matsuyama A, Yoshida M. Yashiroda Y, et al. Curr Opin Chem Biol. 2008 Feb;12(1):55-9. doi: 10.1016/j.cbpa.2008.01.024. Epub 2008 Mar 7. Curr Opin Chem Biol. 2008. PMID: 18282487 Review.
Cited by
- Systematic identification of signaling pathways with potential to confer anticancer drug resistance.
Martz CA, Ottina KA, Singleton KR, Jasper JS, Wardell SE, Peraza-Penton A, Anderson GR, Winter PS, Wang T, Alley HM, Kwong LN, Cooper ZA, Tetzlaff M, Chen PL, Rathmell JC, Flaherty KT, Wargo JA, McDonnell DP, Sabatini DM, Wood KC. Martz CA, et al. Sci Signal. 2014 Dec 23;7(357):ra121. doi: 10.1126/scisignal.aaa1877. Sci Signal. 2014. PMID: 25538079 Free PMC article. - Narrowing the focus: a toolkit to systematically connect oncogenic signaling pathways with cancer phenotypes.
Singleton KR, Wood KC. Singleton KR, et al. Genes Cancer. 2016 Jul;7(7-8):218-228. doi: 10.18632/genesandcancer.112. Genes Cancer. 2016. PMID: 27738492 Free PMC article. Review. - Modeling the complete kinetics of coxsackievirus B3 reveals human determinants of host-cell feedback.
Lopacinski AB, Sweatt AJ, Smolko CM, Gray-Gaillard E, Borgman CA, Shah M, Janes KA. Lopacinski AB, et al. Cell Syst. 2021 Apr 21;12(4):304-323.e13. doi: 10.1016/j.cels.2021.02.004. Epub 2021 Mar 18. Cell Syst. 2021. PMID: 33740397 Free PMC article. - Pooled analysis of radiation hybrids identifies loci for growth and drug action in mammalian cells.
Khan AH, Lin A, Wang RT, Bloom JS, Lange K, Smith DJ. Khan AH, et al. Genome Res. 2020 Oct;30(10):1458-1467. doi: 10.1101/gr.262204.120. Epub 2020 Sep 2. Genome Res. 2020. PMID: 32878976 Free PMC article. - TMEM2 Is a SOX4-Regulated Gene That Mediates Metastatic Migration and Invasion in Breast Cancer.
Lee H, Goodarzi H, Tavazoie SF, Alarcón CR. Lee H, et al. Cancer Res. 2016 Sep 1;76(17):4994-5005. doi: 10.1158/0008-5472.CAN-15-2322. Epub 2016 Jun 21. Cancer Res. 2016. PMID: 27328729 Free PMC article.
References
- Meyerson M, Gabriel S, Getz G. Nat Rev Genet. 2010;11:685–696. - PubMed
- 1000 Genomes Project Consortium et al. Nature. 2010;467:1061–1073. - PubMed
- Moody SE, Boehm JS, Barbie DA, Hahn WC. Cur Opin Mol Ther. 2010;12:284–293. - PubMed
Publication types
MeSH terms
Grants and funding
- R33 CA128625/CA/NCI NIH HHS/United States
- P01 CA142536/CA/NCI NIH HHS/United States
- R33 CA128625-01A1/CA/NCI NIH HHS/United States
- R33 CA128625-02/CA/NCI NIH HHS/United States
- R33 CA128625-03/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials