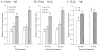Functional activity of the novel Alzheimer's amyloid β-peptide interacting domain (AβID) in the APP and BACE1 promoter sequences and implications in activating apoptotic genes and in amyloidogenesis - PubMed (original) (raw)
Functional activity of the novel Alzheimer's amyloid β-peptide interacting domain (AβID) in the APP and BACE1 promoter sequences and implications in activating apoptotic genes and in amyloidogenesis
Jason A Bailey et al. Gene. 2011.
Abstract
Amyloid-β peptide (Aβ) plaque in the brain is the primary (post mortem) diagnostic criterion of Alzheimer's disease (AD). The physiological role(s) of Aβ are poorly understood. We have previously determined an Aβ interacting domain (AβID) in the promoters of AD-associated genes (Maloney and Lahiri, 2011. Gene. 15,doi:10.1016/j.gene.2011.06.004. epub ahead of print.). This AβID interacts in a DNA sequence-specific manner with Aβ. We now demonstrate novel Aβ activity as a possible transcription factor. Herein, we detected Aβ-chromatin interaction in cell culture by ChIP assay. We observed that human neuroblastoma (SK-N-SH) cells treated with FITC conjugated Aβ1-40 localized Aβ to the nucleus in the presence of H2O2-mediated oxidative stress. Furthermore, primary rat fetal cerebrocortical cultures were transfected with APP and BACE1 promoter-luciferase fusions, and rat PC12 cultures were transfected with polymorphic APP promoter-CAT fusion clones. Transfected cells were treated with different Aβ peptides and/or H2O2. Aβ treatment of cell cultures produced a DNA sequence-specific response in cells transfected with polymorphic APP clones. Our results suggest the Aβ peptide may regulate its own production through feedback on its precursor protein and BACE1, leading to amyloidogenesis in AD.
Copyright © 2011 Elsevier B.V. All rights reserved.
Figures
Fig. 1. Chromatin Immunoprecipitation (ChIP) assay of Aβ in NB cell nuclei
ChIP and PCR were carried out as described in the text. A. ChIP for Aβ binding a 200 bp region containing the predicted APP −3833 AβID. B. ChIP for Aβ binding a 200 bp region containing the BACE1 −119 AβID. C. Precipitated, cross–linked chromatin was probed for presence of Aβ peptide (lanes 4, 8), SP1 (lanes 5, 9), and the N–terminus of the APP protein (lane 6) within the indicated region of the APP promoter. D. Precipitated, cross–linked chromatin was probed for presence of Aβ peptide (lanes 4, 9), SP1 (lanes 5, 10), and β–galactosidase (lanes 6, 11) within the indicated region of the BACE1 promoter. Treatment conditions in the cells and other appropriate controls in the assay were as indicated in the figure.
Fig. 2. Treatment of SK–N–SH cells with Aβ in the presence or absence of H2O2
Human SK–N–SH neuroblastoma cells were treated with FITC–Aβ1–40 in presence or absence of H2O2 as described in the text. Cells were incubated with these treatments for 48 hours and visualized with Hoechst 33342 dye, and EtHD, corresponding to all cell nuclei and dead nuclei, respectively. Cells were fluorescently imaged at appropriate wavelengths for FITC, Hoechst treatment, and EtHD treatment. A and E. with all nuclei labeled with Hoechst 33342. B and F. FITC–Aβ1–40 labeled nuclei. C and G. Dead cells labeled with EtHD. D and G. Composite of all three fluorescent signals overlaid on a phase contrast image of the cells.
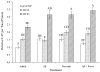
Fig. 3. Effects of Aβ treatment on APP and BACE1 5′–flanking sequence activity
A. A 1.2 kb fragment of the APP 5′–flanking region (Song and Lahiri, 1998) was cloned into the pGL3 luciferase expression vector as described in the text. The APP fragment contains a putative site for Aβ binding. B. A 3.3 kb fragment of the human BACE1 5′–flanking region (Ge et al., 2004b) was cloned into the pGL3 luciferase expression vector as described in the text. The BACE1 fragment contains a confirmed site for Aβ binding, determined herein. Primary rat cerebrocortical neuronal (PRCN) cultures were transiently transfected with a Renilla luciferase control vector and C. APP/pGL3 firefly luciferase fusion clone or D. BACE1/pGL3 luciferase fusion clone. Transfected cells were left untreated or treated with Aβ1–28, 1–40, or 1–42, all 1 μM. Cells were harvested and activities of firefly and Renilla luciferases were measured. Firefly luciferase activity within each treatment was normalized to corresponding Renilla luciferase activity. All within each cell type was then normalized to the average firefly/Renilla ratio for untreated APP/luciferase fusion activity. Letters above data bars indicate general linear mixed model multiple range categories. Samples sharing a letter do not significantly differ at p < 0.05. Thus, no differences in APP promoter activity were found between any treatment group, while there were significant increases in BACE1 promoter activity in the Aβ1–28 and Aβ1–42 treatment groups relative to untreated cells and Aβ1–40 was similar to vehicle.
Fig. 4. Differential response of a single–nucleotide polymorphism of the APP promoter to Aβ treatment, all three effects combined
Polymorphic promoter/enhancer CAT expression clones and pCAT3P were transfected into PC12 cells and the transfected cells were treated with H2O2 and Aβ, singly or in combination, as described in the main text. Reporter CAT ELISA signal was normalized to total cell protein. Results are presented as proportional to untreated pCAT3P vector transfected cells. Letters above data bars indicate general linear mixed model multiple range categories. Statistical significance is indicated by letters above the data bars, with samples sharing a letter do not significantly differ at p < 0.05, and conversely, samples not sharing a letter are significantly different at p < 0.05.
Fig. 5. Differential response of a single–nucleotide polymorphism of the APP promoter to Aβ treatment taken as single effects
Polymorphic promoter/enhancer CAT expression clones and pCAT3P vector plasmid were transfected into PC12 cells and the transfected cells were treated with H2O2 and Aβ, singly or in combination, as described in the main text. Reporter CAT ELISA signal was normalized to total cell protein. A. Both of the clones tested (A and G) had significantly higher activity than the vector, and the G allele was significantly more active than the A allele. B and C. Both Aβ and H2O2 treatment significantly increased activity of this site, independent of the polymorphism. Results are presented as proportional to untreated pCAT3P vector transfected cells. Letters above data bars indicate generalized linear mixed model multiple range categories. Samples sharing a letter do not significantly differ at p < 0.05. For Aβ treatment and H2O2, “*” indicates significant difference at p < 0.05.
Fig. 6. Differential response of a single–nucleotide polymorphism of the APP promoter to Aβ treatment analyzed as pairs of effects
Polymorphic promoter/enhancer CAT expression clones and pCAT3P were transfected into PC12 cells and the transfected cells were treated with H2O2 and Aβ, singly or in combination, as described in the main text. CAT ELISA signal was normalized to total cell protein. Results were then normalized to adjusted signal for untreated pCAT3P transfected cells and expressed as marginal means estimated by holding a single effect (clone, Aβ treatment, or H2O2 treatment) constant. A. Aβ treatment increased transcriptional activity of the G but not the A clone. Both clones were significantly more active than the pCAT3P vector. B. H2O2 treatment increased the activity of both clones, indicating that this effect is not sequence–specific. C. Aβ treatment in the absence of H2O2 increased promoter activity, and H2O2 increased promoter activity significantly both in the presence or absence of Aβ. Letters above data bars indicate generalized linear mixed model multiple range categories. Samples sharing a letter do not significantly differ at p < 0.05. Although the −H2O2/−Aβ treatment had a significant difference from combinations that were treated with H2O2,, Aβ, or H2O2 + Aβ, when examining Aβ × H2O2, the interaction was not significant overall by Type III test.
Fig. 7. Models of Aβ feedback and activity as a transcription factor
A. Non–cytotoxic pathway. Aβ levels are insufficient to significantly activate apoptotic genes such as TP53 and the “cell death pair response” of upregulated ASCL1 and downregulated OLIG2 through their AβID sequences. These levels maintain APP and BACE1 at non–pathogenic levels. B. Cytotoxic pathway. Aβ levels and resulting APP and BACE1 production are stimulated by conditions such as metal–induced oxidation (ROS), which ultimately results in increased production of Aβ peptides. The levels of Aβ are sufficient to significantly increase levels of p53 and induce the “cell death pair response” of upregulated ASCL1 and downregulated OLIG2. Additional Aβ is produced beyond what is required for physiological function, and the excess Aβ becomes deposited as amyloid plaque.
Similar articles
- miR-15b reduces amyloid-β accumulation in SH-SY5Y cell line through targetting NF-κB signaling and BACE1.
Li J, Wang H. Li J, et al. Biosci Rep. 2018 Nov 13;38(6):BSR20180051. doi: 10.1042/BSR20180051. Print 2018 Dec 21. Biosci Rep. 2018. PMID: 29961672 Free PMC article. - Alternative Selection of β-Site APP-Cleaving Enzyme 1 (BACE1) Cleavage Sites in Amyloid β-Protein Precursor (APP) Harboring Protective and Pathogenic Mutations within the Aβ Sequence.
Kimura A, Hata S, Suzuki T. Kimura A, et al. J Biol Chem. 2016 Nov 11;291(46):24041-24053. doi: 10.1074/jbc.M116.744722. Epub 2016 Sep 29. J Biol Chem. 2016. PMID: 27687728 Free PMC article. - The beta-secretase, BACE: a prime drug target for Alzheimer's disease.
Vassar R. Vassar R. J Mol Neurosci. 2001 Oct;17(2):157-70. doi: 10.1385/JMN:17:2:157. J Mol Neurosci. 2001. PMID: 11816789 Review. - BACE1: the beta-secretase enzyme in Alzheimer's disease.
Vassar R. Vassar R. J Mol Neurosci. 2004;23(1-2):105-14. doi: 10.1385/JMN:23:1-2:105. J Mol Neurosci. 2004. PMID: 15126696 Review.
Cited by
- Aberrant Co-localization of Synaptic Proteins Promoted by Alzheimer's Disease Amyloid-β Peptides: Protective Effect of Human Serum Albumin.
Domínguez-Prieto M, Velasco A, Vega L, Tabernero A, Medina JM. Domínguez-Prieto M, et al. J Alzheimers Dis. 2017;55(1):171-182. doi: 10.3233/JAD-160346. J Alzheimers Dis. 2017. PMID: 27662292 Free PMC article. - The Alzheimer's amyloid-degrading peptidase, neprilysin: can we control it?
Nalivaeva NN, Belyaev ND, Zhuravin IA, Turner AJ. Nalivaeva NN, et al. Int J Alzheimers Dis. 2012;2012:383796. doi: 10.1155/2012/383796. Epub 2012 Jul 26. Int J Alzheimers Dis. 2012. PMID: 22900228 Free PMC article. - Tyrosine Rotamer States in Beta Amyloid: Signatures of Aggregation and Fibrillation.
Mancini O, Rolinski OJ, Kubiak-Ossowska K, Mulheran PA. Mancini O, et al. ACS Omega. 2018 Nov 27;3(11):16046-16056. doi: 10.1021/acsomega.8b02408. eCollection 2018 Nov 30. ACS Omega. 2018. PMID: 31458243 Free PMC article. - Powering Amyloid Beta Degrading Enzymes: A Possible Therapy for Alzheimer's Disease.
Sikanyika NL, Parkington HC, Smith AI, Kuruppu S. Sikanyika NL, et al. Neurochem Res. 2019 Jun;44(6):1289-1296. doi: 10.1007/s11064-019-02756-x. Epub 2019 Feb 26. Neurochem Res. 2019. PMID: 30806879 Review. - Selective acetyl- and butyrylcholinesterase inhibitors reduce amyloid-β ex vivo activation of peripheral chemo-cytokines from Alzheimer's disease subjects: exploring the cholinergic anti-inflammatory pathway.
Reale M, Di Nicola M, Velluto L, D'Angelo C, Costantini E, Lahiri DK, Kamal MA, Yu QS, Greig NH. Reale M, et al. Curr Alzheimer Res. 2014;11(6):608-22. doi: 10.2174/1567205010666131212113218. Curr Alzheimer Res. 2014. PMID: 24359497 Free PMC article.
References
- Bailey JA, Lahiri DK. Neuronal differentiation is accompanied by increased levels of SNAP-25 protein in fetal rat primary cortical neurons: implications in neuronal plasticity and Alzheimer’s disease. Ann N Y Acad Sci. 2006;1086:54–65. - PubMed
- Barrantes A, Rejas MT, Benitez MJ, Jimenez JS. Interaction between Alzheimer’s Abeta1–42 peptide and DNA detected by surface plasmon resonance. J Alzheimers Dis. 2007;12:345–55. - PubMed
- Baruch-Suchodolsky R, Fischer B. Abeta40, either soluble or aggregated, is a remarkably potent antioxidant in cell-free oxidative systems. Biochemistry. 2009;48:4354–70. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AG018884/AG/NIA NIH HHS/United States
- R01 AG018379-09/AG/NIA NIH HHS/United States
- R01 AG018379-10S1/AG/NIA NIH HHS/United States
- R01 AG018884-10/AG/NIA NIH HHS/United States
- R21 AG042804/AG/NIA NIH HHS/United States
- R01 AG018379/AG/NIA NIH HHS/United States
- AG18379/AG/NIA NIH HHS/United States
- R01 AG018884-09/AG/NIA NIH HHS/United States
- AG18884/AG/NIA NIH HHS/United States
- R01 AG018379-10/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous