phenosim--A software to simulate phenotypes for testing in genome-wide association studies - PubMed (original) (raw)
phenosim--A software to simulate phenotypes for testing in genome-wide association studies
Torsten Günther et al. BMC Bioinformatics. 2011.
Abstract
Background: There is a great interest in understanding the genetic architecture of complex traits in natural populations. Genome-wide association studies (GWAS) are becoming routine in human, animal and plant genetics to understand the connection between naturally occurring genotypic and phenotypic variation. Coalescent simulations are commonly used in population genetics to simulate genotypes under different parameters and demographic models.
Results: Here, we present phenosim, a software to add a phenotype to genotypes generated in time-efficient coalescent simulations. Both qualitative and quantitative phenotypes can be generated and it is possible to partition phenotypic variation between additive effects and epistatic interactions between causal variants. The output formats of phenosim are directly usable as input for different GWAS tools. The applicability of phenosim is shown by simulating a genome-wide association study in Arabidopsis thaliana.
Conclusions: By using the coalescent approach to generate genotypes and phenosim to add phenotypes, the data sets can be used to assess the influence of various factors such as demography, genetic architecture or selection on the statistical power of association methods to detect causal genetic variants under a wide variety of population genetic scenarios. phenosim is freely available from the authors' website http://evoplant.uni-hohenheim.de.
Figures
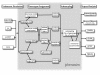
Figure 1
Work flow. Flowchart of the phenosim pipeline.
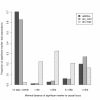
Figure 2
Example. Proportion of significant marker-trait associations (Bonferroni adjusted significance threshold) found at different distances from the causal marker. For each QTN, the distance to the next significant association is shown. The bars on the left show the proportion of simulated data sets for which no significant association was found. The simulated models include: 'additive' - 2 QTNs randomly distributed with a variance proportion of 0.05 each; 'epi_rand' - 2 QTNs randomly distributed with a variance proportion of 0.01 each and 0.08 epistatic effect; 'epi_hapl' - 2 QTNs located on a common haploblock with a variance proportion of 0.01 each and 0.08 epistatic effect. Each model was simulated 1000 times.
Similar articles
- simplePHENOTYPES: SIMulation of pleiotropic, linked and epistatic phenotypes.
Fernandes SB, Lipka AE. Fernandes SB, et al. BMC Bioinformatics. 2020 Oct 31;21(1):491. doi: 10.1186/s12859-020-03804-y. BMC Bioinformatics. 2020. PMID: 33129253 Free PMC article. - GPOPSIM: a simulation tool for whole-genome genetic data.
Zhang Z, Li X, Ding X, Li J, Zhang Q. Zhang Z, et al. BMC Genet. 2015 Feb 5;16(1):10. doi: 10.1186/s12863-015-0173-4. BMC Genet. 2015. PMID: 25652552 Free PMC article. - Marker-based estimation of heritability in immortal populations.
Kruijer W, Boer MP, Malosetti M, Flood PJ, Engel B, Kooke R, Keurentjes JJ, van Eeuwijk FA. Kruijer W, et al. Genetics. 2015 Feb;199(2):379-98. doi: 10.1534/genetics.114.167916. Epub 2014 Dec 19. Genetics. 2015. PMID: 25527288 Free PMC article. - Software engineering the mixed model for genome-wide association studies on large samples.
Zhang Z, Buckler ES, Casstevens TM, Bradbury PJ. Zhang Z, et al. Brief Bioinform. 2009 Nov;10(6):664-75. doi: 10.1093/bib/bbp050. Brief Bioinform. 2009. PMID: 19933212 Review. - An overview of population genetic data simulation.
Yuan X, Miller DJ, Zhang J, Herrington D, Wang Y. Yuan X, et al. J Comput Biol. 2012 Jan;19(1):42-54. doi: 10.1089/cmb.2010.0188. Epub 2011 Dec 9. J Comput Biol. 2012. PMID: 22149682 Free PMC article. Review.
Cited by
- H3AGWAS: a portable workflow for genome wide association studies.
Brandenburg JT, Clark L, Botha G, Panji S, Baichoo S, Fields C, Hazelhurst S. Brandenburg JT, et al. BMC Bioinformatics. 2022 Nov 19;23(1):498. doi: 10.1186/s12859-022-05034-w. BMC Bioinformatics. 2022. PMID: 36402955 Free PMC article. - Identifying Cancer genes by combining two-rounds RWR based on multiple biological data.
Zhang W, Lei Ieee Member X, Bian C. Zhang W, et al. BMC Bioinformatics. 2019 Nov 25;20(Suppl 18):518. doi: 10.1186/s12859-019-3123-8. BMC Bioinformatics. 2019. PMID: 31760937 Free PMC article. - PhenotypeSimulator: A comprehensive framework for simulating multi-trait, multi-locus genotype to phenotype relationships.
Meyer HV, Birney E. Meyer HV, et al. Bioinformatics. 2018 Sep 1;34(17):2951-2956. doi: 10.1093/bioinformatics/bty197. Bioinformatics. 2018. PMID: 29617944 Free PMC article. - Simulating variance heterogeneity in quantitative genome wide association studies.
Al Kawam A, Alshawaqfeh M, Cai JJ, Serpedin E, Datta A. Al Kawam A, et al. BMC Bioinformatics. 2018 Mar 21;19(Suppl 3):72. doi: 10.1186/s12859-018-2061-1. BMC Bioinformatics. 2018. PMID: 29589560 Free PMC article. - cophesim: a comprehensive phenotype simulator for testing novel association methods.
Zhbannikov IY, Arbeev KG, Yashin AI. Zhbannikov IY, et al. F1000Res. 2017 Aug 1;6:1294. doi: 10.12688/f1000research.11968.1. eCollection 2017. F1000Res. 2017. PMID: 28979766 Free PMC article.
References
- Hindorff La, Sethupathy P, Junkins Ha, Ramos EM, Mehta JP, Collins FS, Manolio TA. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(23):9362–7. doi: 10.1073/pnas.0903103106. - DOI - PMC - PubMed
- Atwell S, Huang YS, Vilhjálmsson BJ, Willems G, Horton M, Li Y, Meng D, Platt A, Tarone AM, Hu TT, Jiang R, Muliyati NW, Zhang X, Amer MA, Baxter I, Brachi B, Chory J, Dean C, Debieu M, de Meaux J, Ecker JR, Faure N, Kniskern JM, Jones JDG, Michael T, Nemri A, Roux F, Salt DE, Tang C, Todesco M, Traw MB, Weigel D, Marjoram P, Borevitz JO, Bergelson J, Nordborg M. Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature. 2010;465(7298):627–31. doi: 10.1038/nature08800. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources