Origin of diderm (Gram-negative) bacteria: antibiotic selection pressure rather than endosymbiosis likely led to the evolution of bacterial cells with two membranes - PubMed (original) (raw)
Origin of diderm (Gram-negative) bacteria: antibiotic selection pressure rather than endosymbiosis likely led to the evolution of bacterial cells with two membranes
Radhey S Gupta. Antonie Van Leeuwenhoek. 2011 Aug.
Abstract
The prokaryotic organisms can be divided into two main groups depending upon whether their cell envelopes contain one membrane (monoderms) or two membranes (diderms). It is important to understand how these and other variations that are observed in the cell envelopes of prokaryotic organisms have originated. In 2009, James Lake proposed that cells with two membranes (primarily Gram-negative bacteria) originated from an ancient endosymbiotic event involving an Actinobacteria and a Clostridia (Lake 2009). However, this Perspective argues that this proposal is based on a number of incorrect assumptions and the data presented in support of this model are also of questionable nature. Thus, there is no reliable evidence to support the endosymbiotic origin of double membrane bacteria. In contrast, many observations suggest that antibiotic selection pressure was an important selective force in prokaryotic evolution and that it likely played a central role in the evolution of diderm (Gram-negative) bacteria. Some bacterial phyla, such as Deinococcus-Thermus, which lack lipopolysaccharide (LPS) and yet contain some characteristics of the diderm bacteria, are postulated as evolutionary intermediates (simple diderms) in the transition between the monoderm bacterial taxa and the bacterial groups that have the archetypal LPS-containing outer cell membrane found in Gram-negative bacteria. It is possible to distinguish the two stages in the evolution of diderm-LPS cells (viz. monoderm bacteria → simple diderms lacking LPS → LPS containing archetypal diderm bacteria) by means of conserved inserts in the Hsp70 and Hsp60 proteins. The insert in the Hsp60 protein also distinguishes the traditional Gram-negative diderm bacterial phyla from atypical taxa of diderm bacteria (viz. Negativicutes, Fusobacteria, Synergistetes and Elusimicrobia). The Gram-negative bacterial phyla with an LPS-diderm cell envelope, as defined by the presence of the Hsp60 insert, are indicated to form a monophyletic clade and no loss of the outer membrane from any species from this group seems to have occurred. This argues against the origin of monoderm prokaryotes from diderm bacteria by loss of outer membrane.
Figures
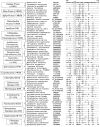
Fig. 1
Partial sequence alignment of the Hsp60 protein showing a 1 aa insert (boxed) in a conserved region that is mainly specific for different bacterial phyla corresponding to traditional Gram-negative bacteria that have an outer cell membrane containing lipopolysaccharide. The presence or absence of this insert in all available sequences from different bacterial groups is indicated along with their names. For example, for Gamma-proteobacteria >500 hits corresponding to Hsp60 were observed and all of them contained this insert (i.e. >500 with insert, 0 without insert). Similarly, for the Actinobacteria phylum, >150 hits were observed and of these only 2 contained the insert (2/>150). Only representative sequences from different bacterial phyla are shown here. The absence of this insert in the Negativicutes, Fusobacteria, Synergistetes and Elusimicrobia distinguishes these atypical diderm taxa from all of the phyla of traditional Gram-negative bacteria that contain this insert. The dashes in the alignment indicate that the same amino acid as that found on the top line (i.e. E. coli protein) is present in that position. The accession numbers of sequences are given in the second column. The numbers on the top indicate the position of this sequence in E. coli protein
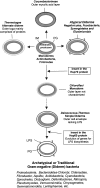
Fig. 2
A cartoon showing the development of outer cell envelopes in various bacterial lineages in response to antibiotic selection pressure (Gupta 2000). The outer cell envelope in Negativicutes, Fusobacteria, Synergistetes and Elusimicrobia (atypical diderm taxa) is distinguished from traditional diderm Gram-negative bacteria by the absence of the Hsp60 insert. The cell membrane from atypical and traditional Gram-negative bacteria are postulated to show significant differences in their biochemical and functional characteristics. The outer cell envelopes of the archetypical Gram-negative phyla are indicated to have evolved from the Chloroflexi and Deinococcus_-Thermus groups of species. Information regarding species distribution of Hsp70 inserts for most bacterial phyla is provided in earlier work (Griffiths and Gupta ; Lake et al._; Singh and Gupta 2009). Abbreviations: PG peptidoglycan, IM inner membrane, LPS lipopolysaccharides
Similar articles
- One or two membranes? Diderm Firmicutes challenge the Gram-positive/Gram-negative divide.
Megrian D, Taib N, Witwinowski J, Beloin C, Gribaldo S. Megrian D, et al. Mol Microbiol. 2020 Mar;113(3):659-671. doi: 10.1111/mmi.14469. Mol Microbiol. 2020. PMID: 31975449 Review. - Photosynthetic Systems Suggest an Evolutionary Pathway to Diderms.
Rogers SO. Rogers SO. Acta Biotheor. 2021 Sep;69(3):343-358. doi: 10.1007/s10441-020-09402-y. Epub 2020 Dec 7. Acta Biotheor. 2021. PMID: 33284411 Free PMC article. - Genome-wide analysis of the Firmicutes illuminates the diderm/monoderm transition.
Taib N, Megrian D, Witwinowski J, Adam P, Poppleton D, Borrel G, Beloin C, Gribaldo S. Taib N, et al. Nat Ecol Evol. 2020 Dec;4(12):1661-1672. doi: 10.1038/s41559-020-01299-7. Epub 2020 Oct 19. Nat Ecol Evol. 2020. PMID: 33077930 - The natural evolutionary relationships among prokaryotes.
Gupta RS. Gupta RS. Crit Rev Microbiol. 2000;26(2):111-31. doi: 10.1080/10408410091154219. Crit Rev Microbiol. 2000. PMID: 10890353 Review.
Cited by
- Complete genome sequence of Acidaminococcus intestini RYC-MR95, a Gram-negative bacterium from the phylum Firmicutes.
D'Auria G, Galán JC, Rodríguez-Alcayna M, Moya A, Baquero F, Latorre A. D'Auria G, et al. J Bacteriol. 2011 Dec;193(24):7008-9. doi: 10.1128/JB.06301-11. J Bacteriol. 2011. PMID: 22123762 Free PMC article. - Investigating the Bactericidal Activity of an Ocular Solution Containing EDTA, Tris, and Polysorbate 80 and Its Impact on the In Vitro Efficacy of Neomycin Sulfate against Staphylococcus aureus: A Preliminary Study.
Amiriantz S, Hoummady S, Jarousse E, Roudeix S, Philippon T. Amiriantz S, et al. Antibiotics (Basel). 2024 Jun 30;13(7):611. doi: 10.3390/antibiotics13070611. Antibiotics (Basel). 2024. PMID: 39061293 Free PMC article. - Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes.
Albertsen M, Hugenholtz P, Skarshewski A, Nielsen KL, Tyson GW, Nielsen PH. Albertsen M, et al. Nat Biotechnol. 2013 Jun;31(6):533-8. doi: 10.1038/nbt.2579. Epub 2013 May 26. Nat Biotechnol. 2013. PMID: 23707974 - The treatment-naive microbiome in new-onset Crohn's disease.
Gevers D, Kugathasan S, Denson LA, Vázquez-Baeza Y, Van Treuren W, Ren B, Schwager E, Knights D, Song SJ, Yassour M, Morgan XC, Kostic AD, Luo C, González A, McDonald D, Haberman Y, Walters T, Baker S, Rosh J, Stephens M, Heyman M, Markowitz J, Baldassano R, Griffiths A, Sylvester F, Mack D, Kim S, Crandall W, Hyams J, Huttenhower C, Knight R, Xavier RJ. Gevers D, et al. Cell Host Microbe. 2014 Mar 12;15(3):382-392. doi: 10.1016/j.chom.2014.02.005. Cell Host Microbe. 2014. PMID: 24629344 Free PMC article. - Veillonella, Firmicutes: Microbes disguised as Gram negatives.
Vesth T, Ozen A, Andersen SC, Kaas RS, Lukjancenko O, Bohlin J, Nookaew I, Wassenaar TM, Ussery DW. Vesth T, et al. Stand Genomic Sci. 2013 Dec 15;9(2):431-48. doi: 10.4056/sigs.2981345. eCollection 2013 Dec 20. Stand Genomic Sci. 2013. PMID: 24976898 Free PMC article.
References
- Cavalier-Smith T. Origins of secondary metabolism. Ciba Found Symp. 1992;171:64–80. - PubMed
- Cavalier-Smith T. The neomuran origin of archaebacteria, the negibacterial root of the universal tree and bacterial megaclassification. Int J Syst Evol Microbiol. 2002;52:7–76. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous