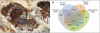The genome of the leaf-cutting ant Acromyrmex echinatior suggests key adaptations to advanced social life and fungus farming - PubMed (original) (raw)
. 2011 Aug;21(8):1339-48.
doi: 10.1101/gr.121392.111. Epub 2011 Jun 30.
Guojie Zhang, Morten Schiøtt, Cai Li, Yannick Wurm, Haofu Hu, Jiajian Zhou, Lu Ji, Feng Qiu, Morten Rasmussen, Hailin Pan, Frank Hauser, Anders Krogh, Cornelis J P Grimmelikhuijzen, Jun Wang, Jacobus J Boomsma
Affiliations
- PMID: 21719571
- PMCID: PMC3149500
- DOI: 10.1101/gr.121392.111
The genome of the leaf-cutting ant Acromyrmex echinatior suggests key adaptations to advanced social life and fungus farming
Sanne Nygaard et al. Genome Res. 2011 Aug.
Abstract
We present a high-quality (>100× depth) Illumina genome sequence of the leaf-cutting ant Acromyrmex echinatior, a model species for symbiosis and reproductive conflict studies. We compare this genome with three previously sequenced genomes of ants from different subfamilies and focus our analyses on aspects of the genome likely to be associated with known evolutionary changes. The first is the specialized fungal diet of A. echinatior, where we find gene loss in the ant's arginine synthesis pathway, loss of detoxification genes, and expansion of a group of peptidase proteins. One of these is a unique ant-derived contribution to the fecal fluid, which otherwise consists of "garden manuring" fungal enzymes that are unaffected by ant digestion. The second is multiple mating of queens and ejaculate competition, which may be associated with a greatly expanded nardilysin-like peptidase gene family. The third is sex determination, where we could identify only a single homolog of the feminizer gene. As other ants and the honeybee have duplications of this gene, we hypothesize that this may partly explain the frequent production of diploid male larvae in A. echinatior. The fourth is the evolution of eusociality, where we find a highly conserved ant-specific profile of neuropeptide genes that may be related to caste determination. These first analyses of the A. echinatior genome indicate that considerable genetic changes are likely to have accompanied the transition from hunter-gathering to agricultural food production 50 million years ago, and the transition from single to multiple queen mating 10 million years ago.
Figures
Figure 1.
The leafcutter ant A. echinatior and annotation of its protein-coding genes. (A) A winged male of the Panamanian leaf-cutting ant A. echinatior in the fungus garden that is maintained by his major- and minor-worker sisters. (B) The total of 17,278 annotated protein-coding genes as obtained from de novo predictions, GLEAN acceptance, homology (to C. floridanus, H. saltator, A. mellifera, N. vitripennis, D. melanogaster, C. elegans, or H. sapiens) and transcriptome evidence. (Photo courtesy of David R. Nash © 2010.)
Figure 2.
Missing genes in the arginine biosynthesis pathway. The specific loss in A. echinatior of two genes that encode enzymes catalyzing two consecutive (final) steps in the biosynthesis of the amino acid arginine. Enzymes are denoted by purple boxes with the EC numbers inside. Pale purple boxes with dashed red borders indicate the two lost or pseudogenized genes.
Figure 3.
Peptidase expansions in the genome of A. echinatior. (A) Expansion of the M16 peptidase gene family with the insulin degrading enzyme, present in one or two copies in all investigated insect genomes (below dotted line), and nardilysin genes (above dotted line). The A. echinatior genes in each group are highlighted in yellow. Bootstrap support values >60% are given. (B) Expansion of the M14 peptidase gene family, with a dotted line separating two subfamilies. The A. echinatior genes are highlighted in yellow. Bootstrap support values >60% are given. Phum was included to increase resolution of this tree. Species (A. echinatior: Aech; H. saltator: Hsal; C. floridanus: Cflo; S. invicta: Sinv; A. mellifera: Amel; D. melanogaster: Dmel; N. vitripennis: Nvit; Pediculus humanus: Phum) and GenBank ID are given for each sequence.
Similar articles
- Leaf-cutting ant fungi produce cell wall degrading pectinase complexes reminiscent of phytopathogenic fungi.
Schiøtt M, Rogowska-Wrzesinska A, Roepstorff P, Boomsma JJ. Schiøtt M, et al. BMC Biol. 2010 Dec 31;8:156. doi: 10.1186/1741-7007-8-156. BMC Biol. 2010. PMID: 21194476 Free PMC article. - Laccase detoxification mediates the nutritional alliance between leaf-cutting ants and fungus-garden symbionts.
De Fine Licht HH, Schiøtt M, Rogowska-Wrzesinska A, Nygaard S, Roepstorff P, Boomsma JJ. De Fine Licht HH, et al. Proc Natl Acad Sci U S A. 2013 Jan 8;110(2):583-7. doi: 10.1073/pnas.1212709110. Epub 2012 Dec 24. Proc Natl Acad Sci U S A. 2013. PMID: 23267060 Free PMC article. - Proteomics reveals synergy between biomass degrading enzymes and inorganic Fenton chemistry in leaf-cutting ant colonies.
Schiøtt M, Boomsma JJ. Schiøtt M, et al. Elife. 2021 Jan 12;10:e61816. doi: 10.7554/eLife.61816. Elife. 2021. PMID: 33433325 Free PMC article. - The prominent role of fungi and fungal enzymes in the ant-fungus biomass conversion symbiosis.
Lange L, Grell MN. Lange L, et al. Appl Microbiol Biotechnol. 2014 Jun;98(11):4839-51. doi: 10.1007/s00253-014-5708-5. Epub 2014 Apr 15. Appl Microbiol Biotechnol. 2014. PMID: 24728757 Review. - The origin of the attine ant-fungus mutualism.
Mueller UG, Schultz TR, Currie CR, Adams RM, Malloch D. Mueller UG, et al. Q Rev Biol. 2001 Jun;76(2):169-97. doi: 10.1086/393867. Q Rev Biol. 2001. PMID: 11409051 Review.
Cited by
- RES-Scanner: a software package for genome-wide identification of RNA-editing sites.
Wang Z, Lian J, Li Q, Zhang P, Zhou Y, Zhan X, Zhang G. Wang Z, et al. Gigascience. 2016 Aug 18;5(1):37. doi: 10.1186/s13742-016-0143-4. Gigascience. 2016. PMID: 27538485 Free PMC article. - ArthropodaCyc: a CycADS powered collection of BioCyc databases to analyse and compare metabolism of arthropods.
Baa-Puyoulet P, Parisot N, Febvay G, Huerta-Cepas J, Vellozo AF, Gabaldón T, Calevro F, Charles H, Colella S. Baa-Puyoulet P, et al. Database (Oxford). 2016 May 30;2016:baw081. doi: 10.1093/database/baw081. Print 2016. Database (Oxford). 2016. PMID: 27242037 Free PMC article. - Amino acid synthesis loss in parasitoid wasps and other hymenopterans.
Ye X, Xiong S, Teng Z, Yang Y, Wang J, Yu K, Wu H, Mei Y, Yan Z, Cheng S, Yin C, Wang F, Yao H, Fang Q, Song Q, Werren JH, Ye G, Li F. Ye X, et al. Elife. 2020 Oct 19;9:e59795. doi: 10.7554/eLife.59795. Elife. 2020. PMID: 33074103 Free PMC article. Retracted. - Relaxed selection underlies genome erosion in socially parasitic ant species.
Schrader L, Pan H, Bollazzi M, Schiøtt M, Larabee FJ, Bi X, Deng Y, Zhang G, Boomsma JJ, Rabeling C. Schrader L, et al. Nat Commun. 2021 May 18;12(1):2918. doi: 10.1038/s41467-021-23178-w. Nat Commun. 2021. PMID: 34006882 Free PMC article. - Hymenoptera Genome Database: new genomes and annotation datasets for improved go enrichment and orthologue analyses.
Walsh AT, Triant DA, Le Tourneau JJ, Shamimuzzaman M, Elsik CG. Walsh AT, et al. Nucleic Acids Res. 2022 Jan 7;50(D1):D1032-D1039. doi: 10.1093/nar/gkab1018. Nucleic Acids Res. 2022. PMID: 34747465 Free PMC article.
References
- Aanen DK, de Fine Licht HH, Debets AJM, Kerstes NAG, Hoekstra RF, Boomsma JJ 2009. High symbiont relatedness stabilizes mutualistic cooperation in fungus-growing termites. Science 326: 1103–1106 - PubMed
- Armitage S, Broch J, Fernández Marín H, Nash D, Boomsma J 2011. Immune defence in leaf-cutting ants: a cross-fostering approach. Evolution 65: 1791–1799 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials