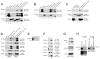Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair - PubMed (original) (raw)
. 2011 Jul 8;146(1):67-79.
doi: 10.1016/j.cell.2011.06.020. Epub 2011 Jun 30.
Jinfei Xu, Mara Sannai, Robert Moore, Elena Caretti, Antonio Cigliano, Madeleine Le Coz, Karthik Devarajan, Andy Wessels, Dianne Soprano, Lara K Abramowitz, Marisa S Bartolomei, Florian Rambow, Maria Rosaria Bassi, Tiziana Bruno, Maurizio Fanciulli, Catherine Renner, Andres J Klein-Szanto, Yoshihiro Matsumoto, Dominique Kobi, Irwin Davidson, Christophe Alberti, Lionel Larue, Alfonso Bellacosa
Affiliations
- PMID: 21722948
- PMCID: PMC3230223
- DOI: 10.1016/j.cell.2011.06.020
Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair
Salvatore Cortellino et al. Cell. 2011.
Abstract
DNA methylation is a major epigenetic mechanism for gene silencing. Whereas methyltransferases mediate cytosine methylation, it is less clear how unmethylated regions in mammalian genomes are protected from de novo methylation and whether an active demethylating activity is involved. Here, we show that either knockout or catalytic inactivation of the DNA repair enzyme thymine DNA glycosylase (TDG) leads to embryonic lethality in mice. TDG is necessary for recruiting p300 to retinoic acid (RA)-regulated promoters, protection of CpG islands from hypermethylation, and active demethylation of tissue-specific developmentally and hormonally regulated promoters and enhancers. TDG interacts with the deaminase AID and the damage response protein GADD45a. These findings highlight a dual role for TDG in promoting proper epigenetic states during development and suggest a two-step mechanism for DNA demethylation in mammals, whereby 5-methylcytosine and 5-hydroxymethylcytosine are first deaminated by AID to thymine and 5-hydroxymethyluracil, respectively, followed by TDG-mediated thymine and 5-hydroxymethyluracil excision repair.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures
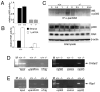
Figure 1. Developmental defects in _Tdg_-null embryos
(A) The expression of Tdg was monitored by western blot. No expression was detected in MEFs homozygous for the _Tdg_- allele, whereas heterozygous MEFs showed reduced expression compared to wild type cells. (B) Tdg is the foremost G:T mismatch repair activity at CpG dinucleotides in MEFs. Repair of a double stranded oligonucleotide containing a G:T mismatch by nuclear extracts of MEFs with different genotypes, in comparison to a substrate neither exposed to lysate nor enzyme (O). Reaction with recombinant MBD4/MED1 (rM) was used as a size marker for cleavage at the mismatched thymine. (C and D) Gross phenotype of wild type and Tdg-/- littermate embryos at embryonic day E11.5: double-arrows show constriction in the cervical region of _Tdg_-null embryos (D) compared to wild type embryos (C); white asterisks mark the carotid artery that is stenotic in _Tdg_-null embryos; the enlarged heart with pericardial effusion (h) and hemorrhagic liver (l) are apparent; arrowheads point to hemorrhagic lesions in the cranium, and enlarged and irregular segmental arteries. (E and F) Cardiac perfusion with India ink in wild type and Tdg-mutant embryos at E11; in _Tdg_-null embryo (F), circulatory insufficiency is demonstrated by reduced perfusion of the dorsal aorta (da) and carotid artery (ca), whereas the third (a3) and fourth (a4) branchial arch arteries are enlarged in comparison to wild type embryos (E); the first (b1) and second (b2) branchial arches, as well as the otic vesicle (ov), are indicated. (G and H) Transverse sections of the liver at E11; compared to wild type (G), the mutant liver has enlarged hepatic sinusoids, the likely proximal causes of abdominal hemorrhage. (I and J) Transverse sections of the heart at E11.5 show a patterning defects in mutant embryos; the conal part of the OFT is severely hyperplastic in mutant embryos (twin arrows in J) when compared to the heterozygous specimen (I), generating an atypical indentation between the right ventricle (RV) and OFT (arrow head in J); the characteristic “dog-leg bend” of the OFT, which is responsible for correctly positioning the OFT over the midline of left ventricle (LV) and RV is not observed in mutant hearts; instead, the OFT is situated right above the RV; as a result, the left part of the body wall is pushed out by the LV (asterisks). (K and L) Immunostaining of the vascular labyrinth with a PECAM/CD31 antibody in wild type (K) and _Tdg_-null (L) embryos at E11 reveal a generalized disorganization of the vascular network in the latter; arrowheads point to irregular branches of the internal carotid with varicosities, bulges and ectasias. See also Figures S1, S2 and S3.
Figure 2. Involvement of TDG in transcription and composition of RAR-p300 complexes
(A) Reduced p300-induced transcriptional activation in _Tdg_-/- MEFs. Luciferase activity of a Gal4 operator luciferase reporter co-transfected with a Gal4 DNA binding domain-p300 fusion construct, and normalized to transfection efficiency using β-galactosidase expression. Since the transfected plasmids are unmethylated, this assay reflects only the co-activator role of TDG. (B) Reduced retinoic-dependent RAR/RXR transcriptional activity in _Tdg_-/- MEFs. CAT activity of a RARE-containing reporter was normalized to transfection efficiency using β-galactosidase expression. (C) Co-IP with an antibody capable of recognizing all the RARs shows lack of association between RAR and p300 in _Tdg_-/- MEFs. Wild type and mutant MEFs were stimulated with 1 μM RA for the indicated time. Approximately equal levels of p300 and RAR are present in wild type and mutant cells. Detection of β-actin is shown as a loading control. (D and E) Chromatin immunoprecipitation shows that Tdg binds directly to the promoter of two differentially expressed, RAR-RXR target genes, Crabp2 (D) and Rbp1 (E), and is required for p300 recruitment and histone H3 acetylation. Approximately equal amounts of input chromatin were used for immunoprecipitation. As negative control, immunoprecipitation with non-specific immunoglobulins was performed. Data are presented as mean ± standard error of the mean (SEM). See also Figure S4.
Figure 3. Hypermethylation of CpG islands in the absence of TDG
(A-E) DNA methylation analysis by sodium bisulfite modification and sequencing of cloned PCR products in wild type and _Tdg_-null cells. Open and closed circles represent unmethylated and methylated CpGs, respectively. Crabp2 (A), Efs (C) and Hoxa5 (D) promoters are unmethylated at various degrees in wild type cells, whereas their CpG islands are hypermethylated in _Tdg_-null MEFs. For the H19 promoter (B), the first three clones in wild type MEFs are likely derivatives of the inactive paternal allele, and the remaining ones are probably of maternal origin (H19 is maternally expressed), whereas in _Tdg_-null MEFs both alleles are hypermethylated. Analysis of the 3′ half of the Igf2 DMR2 (E): the first five clones in wild type PGCs are likely derived from the paternal (methylated) allele, and the remaining ones are probably of maternal origin, whereas in _Tdg_-null PGCs, all the alleles are hypermethylated. See also Figure S4.
Figure 4. TDG is involved in DNA demethylation
(A) DNA methylation analysis of five CpG dinucleotides of the Alb1 enhancer in liver and brain of wild type and _Tdg_-null embryos at E11; the numbers refer to the position of CpGs relative to the transcription start site (TSS). (B) Corresponding quantification and color-coded display of percent DNA methylation at each CpG dinucleotide of the Alb1 enhancer. (C) Real-time RT-PCR quantification of Alb1 mRNA expression, normalized to Hprt mRNA expression, in wild type and _Tdg_-null livers at E11. (D) Methylated DNA immunoprecipitation-quantitative PCR (MeDIP-qPCR) analysis of methylation levels at the Tat gene GRU, expressed as percent immunoprecipitated DNA relative to input DNA, in ES cells, wild type and _Tdg_-null embryos at E10.5 (headless embryo body dissected to enrich for liver). (E) Methylation analysis by sodium bisulfite modification and sequencing of five CpG dinucleotides of the Tat gene GRU (at -2520, -2485, -2473, -2390, and -2386 bp relative to TSS) in ES cells, wild type, and _Tdg_-null embryos at E10.5 (dissected to enrich for liver); two-sided Fisher's Exact Test at the 5% significance level. (F) Western blot analysis showing effective downregulation of TDG in P19 C8 cells expressing a short hairpin RNA (shRNA) directed against murine Tdg mRNA in comparison to parental P19 cells and control shRNA P19 C7 cells. (G) Detection by fluorescence of GFP+ cells (top) in cultures of parental P19, TDG-shRNA-containing P19 C8 cells and control shRNA C7 cells, transfected with unmethylated and SssI-methylated human Oct4_∷_EGFP reporter. Cells were plated at approximately equal density, as evidenced by phase contrast microscopy (bottom). (H) Quantitation of expression of unmethylated and SssI-methylated human Oct4_∷_EGFP reporter in P19, TDG-shRNA C8 and control shRNA C7 cells. (I) DNA methylation analysis by sodium bisulfite modification and sequencing of the proximal region (region 8)(Deb-Rinker et al., 2005) of the human Oct4 promoter from the untransfected methylated Oct4_∷_EGFP reporter and the same reporter transfected and recovered from P19 and C8 cells; two-sided Fisher's Exact Test at the 5% significance level. Data are presented as mean ± standard error of the mean (SEM).
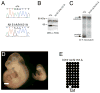
Figure 5. The DNA glycosylase activity of TDG is required for development and DNA demethylation
(A) Sequence analysis of a cDNA fragment encompassing the relevant Tdg exon 4 region from wild type and _Tdg_N151A/N151A E10.5 embryo total RNA confirms expression of the knock-in allele. (B) Western blot analysis with an anti-TDG antibody reveals expression of the wild type and TDGN151A protein in E10.5 embryo lysates of corresponding genotypes. (C) Repair of a double stranded oligonucleotide containing a G:T mismatch by nuclear extracts of whole E10.5 embryo extracts of the indicated genotypes. (D) Gross phenotype of wild type (left) and TdgN151A/N151A (right) littermate embryos at embryonic day E10.5. Size bar corresponds to 750 μm. (E) Methylation analysis by sodium bisulfite modification and sequencing of five CpG dinucleotides of the Tat gene GRU in a _Tdg_N151A/N151A E10.5 embryo (headless embryo body preparations dissected to enrich for liver); comparison of methylation levels with the wild type embryo in Figure 4E was made using the two-sided Fisher's Exact Test at the 5% significance level and revealed a _p_-value equal to 0.0004. See also Figure S1.
Figure 6. TDG is in a complex with AID and GADD45a
(A-D) Immunoprecipitates of lysates of HEK-293 (A-C) or A549 and U251 (D) cells transfected with hemagglutinin (HA)-, FLAG- or MYC-tagged expression constructs were resolved by PAGE and detected by western blotting with the indicated antibodies. Western blotting of lysates shows that identical tagged constructs were expressed at approximately equal levels. In these and other Western blots, the presence of additional TDG bands is the result of sumoylation or other post-translational modifications (Cortazar et al., 2007). Note that in (C) FLAG-tagged AID and FLAG-tagged GADD45a immunoprecipitate not only with exogenous, HA-tagged TDG (lanes 6 and 8) but also with endogenous TDG (lanes 3 and 5). (E) Western blotting with an anti-TDG antibody reveals TDG expression in HEK-293 cells but not in A549 or U251 cells. (F) Co-immunoprecipitation experiments with the indicated antibodies show that AID forms a complex with TDG and GADD45a at endogenous levels of expression in P19 embryonic carcinoma cells. A western with anti-AID antibody confirms that AID was actually immunoprecipitated. (G) AID levels are reduced by shRNA in the P19 derivative, TDG knockdown cell line C8, as evidenced by western blotting of lysates with the indicated anti-TDG and anti-AID antibodies; western blotting with an anti-β-actin antibody acts as a loading control. (H) The indicated recombinant proteins were pre-mixed and the mixtures were immunoprecipitated with an anti-AID antibody; immunoprecipitates along with the input recombinant TDG or GADD45a were detected by western blotting with an anti-TDG or anti-GADD45a antibody, as indicated. GADD45a is readily detected, whereas only a small amount of TDG (visible in the longer exposure, bottom panel, left) precipitates with AID, suggesting a low-affinity interaction. See also Figures S5 and S6.
Figure 7. TDG glycosylase activity on 5hmU and model of the role of TDG in DNA demethylation pathways
(A) Recombinant TDG and related glycosylases were incubated with 5hmU-containing single-strand oligonucleotide or double-strand oligonucleotides bearing 5hmU:A pairing or 5hmU:G mismatch, all 32P-labeled on the 5hmU strand. The resulting AP site was cleaved with alkali at high temperature. (B) Schematic of the involvement of TDG in both the deamination and hydroxylation-deamination pathways of DNA demethylation. Lethality/viability of the glycosylase knock-out mice is indicated. See also Figures S5 and S7.
Similar articles
- Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: potential implications for active demethylation of CpG sites.
Maiti A, Drohat AC. Maiti A, et al. J Biol Chem. 2011 Oct 14;286(41):35334-35338. doi: 10.1074/jbc.C111.284620. Epub 2011 Aug 23. J Biol Chem. 2011. PMID: 21862836 Free PMC article. - Genome-wide distribution of 5-formylcytosine in embryonic stem cells is associated with transcription and depends on thymine DNA glycosylase.
Raiber EA, Beraldi D, Ficz G, Burgess HE, Branco MR, Murat P, Oxley D, Booth MJ, Reik W, Balasubramanian S. Raiber EA, et al. Genome Biol. 2012 Aug 17;13(8):R69. doi: 10.1186/gb-2012-13-8-r69. Genome Biol. 2012. PMID: 22902005 Free PMC article. - Embryonic lethal phenotype reveals a function of TDG in maintaining epigenetic stability.
Cortázar D, Kunz C, Selfridge J, Lettieri T, Saito Y, MacDougall E, Wirz A, Schuermann D, Jacobs AL, Siegrist F, Steinacher R, Jiricny J, Bird A, Schär P. Cortázar D, et al. Nature. 2011 Feb 17;470(7334):419-23. doi: 10.1038/nature09672. Epub 2011 Jan 30. Nature. 2011. PMID: 21278727 - Epigenetic modifications in DNA could mimic oxidative DNA damage: A double-edged sword.
Ito S, Kuraoka I. Ito S, et al. DNA Repair (Amst). 2015 Aug;32:52-57. doi: 10.1016/j.dnarep.2015.04.013. Epub 2015 May 1. DNA Repair (Amst). 2015. PMID: 25956859 Review. - Role of base excision repair in maintaining the genetic and epigenetic integrity of CpG sites.
Bellacosa A, Drohat AC. Bellacosa A, et al. DNA Repair (Amst). 2015 Aug;32:33-42. doi: 10.1016/j.dnarep.2015.04.011. Epub 2015 May 1. DNA Repair (Amst). 2015. PMID: 26021671 Free PMC article. Review.
Cited by
- Biochemical and structural characterization of the glycosylase domain of MBD4 bound to thymine and 5-hydroxymethyuracil-containing DNA.
Moréra S, Grin I, Vigouroux A, Couvé S, Henriot V, Saparbaev M, Ishchenko AA. Moréra S, et al. Nucleic Acids Res. 2012 Oct;40(19):9917-26. doi: 10.1093/nar/gks714. Epub 2012 Jul 30. Nucleic Acids Res. 2012. PMID: 22848106 Free PMC article. - Mitochondrial membrane potential regulates nuclear DNA methylation and gene expression through phospholipid remodeling.
Mori MP, Lozoya O, Brooks AM, Grenet D, Nadalutti CA, Ryback B, Huang KT, Hasan P, Hajnóczky G, Santos JH. Mori MP, et al. bioRxiv [Preprint]. 2024 Jan 13:2024.01.12.575075. doi: 10.1101/2024.01.12.575075. bioRxiv. 2024. PMID: 38260521 Free PMC article. Updated. Preprint. - DNA methylation in schizophrenia: progress and challenges of epigenetic studies.
Nishioka M, Bundo M, Kasai K, Iwamoto K. Nishioka M, et al. Genome Med. 2012 Dec 13;4(12):96. doi: 10.1186/gm397. eCollection 2012. Genome Med. 2012. PMID: 23234572 Free PMC article. Review. - Dose dependent effects on cell cycle checkpoints and DNA repair by bendamustine.
Beeharry N, Rattner JB, Bellacosa A, Smith MR, Yen TJ. Beeharry N, et al. PLoS One. 2012;7(6):e40342. doi: 10.1371/journal.pone.0040342. Epub 2012 Jun 29. PLoS One. 2012. PMID: 22768280 Free PMC article. - Engineered deaminases as a key component of DNA and RNA editing tools.
Budzko L, Hoffa-Sobiech K, Jackowiak P, Figlerowicz M. Budzko L, et al. Mol Ther Nucleic Acids. 2023 Oct 20;34:102062. doi: 10.1016/j.omtn.2023.102062. eCollection 2023 Dec 12. Mol Ther Nucleic Acids. 2023. PMID: 38028200 Free PMC article. Review.
References
- Baker D, Liu P, Burdzy A, Sowers LC. Characterization of the substrate specificity of a human 5-hydroxymethyluracil glycosylase activity. Chem Res Toxicol. 2002;15:33–39. - PubMed
- Barreto G, Schafer A, Marhold J, Stach D, Swaminathan SK, Handa V, Doderlein G, Maltry N, Wu W, Lyko F, et al. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature. 2007;445:671–675. - PubMed
- Bird A. The essentials of DNA methylation. Cell. 1992;70:5–8. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- DK067558/DK/NIDDK NIH HHS/United States
- CA06927/CA/NCI NIH HHS/United States
- T32 GM008216/GM/NIGMS NIH HHS/United States
- R29 CA078412/CA/NCI NIH HHS/United States
- CA78412/CA/NCI NIH HHS/United States
- P30 CA006927/CA/NCI NIH HHS/United States
- T32GM008216/GM/NIGMS NIH HHS/United States
- R01 CA078412/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous