Why do RNA viruses recombine? - PubMed (original) (raw)
Review
Why do RNA viruses recombine?
Etienne Simon-Loriere et al. Nat Rev Microbiol. 2011.
Abstract
Recombination occurs in many RNA viruses and can be of major evolutionary significance. However, rates of recombination vary dramatically among RNA viruses, which can range from clonal to highly recombinogenic. Here, we review the factors that might explain this variation in recombination frequency and show that there is little evidence that recombination is favoured by natural selection to create advantageous genotypes or purge deleterious mutations, as predicted if recombination functions as a form of sexual reproduction. Rather, recombination rates seemingly reflect larger-scale patterns of viral genome organization, such that recombination may be a mechanistic by-product of the evolutionary pressures acting on other aspects of virus biology.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
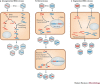
Figure 1. Generation of recombinant and reassortant RNA viruses.
a | Co-infection of a cell by genetically distinct viral strains can lead to the generation of recombinant viruses. This process can occur in both non-segmented viruses (as shown here) or within a segment of a segmented virus. b | Co-infection of a cell by genetically distinct strains of a retrovirus can lead to the generation of 'heterozygous' virus particles, after which a template-switching event can lead to a recombinant provirus. c | Co-infection of a cell by genetically distinct strains of a segmented virus can generate different combinations of reassortant progeny.
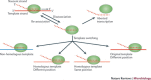
Figure 2. Potential consequences of a disassociation event during viral transcription.
Following disassociation of the viral polymerase and the nascent nucleic acid from the template, the polymerase has to find a template or the transcription process will abort. This re-association event can take place on the same template (red), at the same genomic position or at a different position. Alternatively, the polymerase can associate with a homologous template (orange), again either at the same genomic position or at a different position. Finally, re-association can take place on a non-homologous template, such as a cellular RNA (blue). Of all these possible RNA recombination events, those occurring at the same position on a homologous template are the most likely to generate functional progeny.
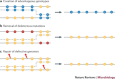
Figure 3. Evolutionary consequences of recombination.
Depending on the acceptor and donor genotypes, and the position of the template switch, recombination can have several positive effects on the genome. Yellow circles indicate wild-type loci. a | Recombination can create advantageous combinations of mutations (blue circles) that increase the rate of adaptive evolution compared with mutation alone, or it can disassociate advantageous and deleterious mutations, allowing the former to spread. b | Recombination can remove deleterious mutations (red circles) and restore the wild-type (fit) genotype, which can lead to a selective advantage for recombination if deleterious mutations occur frequently enough and interact synergistically. c | Recombination can also generate a functional genome from damaged parental molecules. Genetic damage, such as strand breaks or oxidative base modifications, are represented by red lightning symbols.
Similar articles
- Phylogenetic analysis reveals a low rate of homologous recombination in negative-sense RNA viruses.
Chare ER, Gould EA, Holmes EC. Chare ER, et al. J Gen Virol. 2003 Oct;84(Pt 10):2691-2703. doi: 10.1099/vir.0.19277-0. J Gen Virol. 2003. PMID: 13679603 - Mechanisms and consequences of positive-strand RNA virus recombination.
Bentley K, Evans DJ. Bentley K, et al. J Gen Virol. 2018 Oct;99(10):1345-1356. doi: 10.1099/jgv.0.001142. Epub 2018 Aug 29. J Gen Virol. 2018. PMID: 30156526 Review. - Recombination, selection and clock-like evolution of Rice yellow mottle virus.
Pinel-Galzi A, Mpunami A, Sangu E, Rakotomalala M, Traoré O, Sérémé D, Sorho F, Séré Y, Kanyeka Z, Konaté G, Fargette D. Pinel-Galzi A, et al. Virology. 2009 Nov 10;394(1):164-72. doi: 10.1016/j.virol.2009.08.008. Epub 2009 Sep 9. Virology. 2009. PMID: 19740507 - [RNA recombination in plant RNA viruses].
Masuda C. Masuda C. Uirusu. 2000 Dec;50(2):233-41. Uirusu. 2000. PMID: 11276812 Review. Japanese. No abstract available. - A phylogenetic survey of recombination frequency in plant RNA viruses.
Chare ER, Holmes EC. Chare ER, et al. Arch Virol. 2006 May;151(5):933-46. doi: 10.1007/s00705-005-0675-x. Epub 2005 Nov 15. Arch Virol. 2006. PMID: 16292597
Cited by
- Determinants of Virus Variation, Evolution, and Host Adaptation.
LaTourrette K, Garcia-Ruiz H. LaTourrette K, et al. Pathogens. 2022 Sep 13;11(9):1039. doi: 10.3390/pathogens11091039. Pathogens. 2022. PMID: 36145471 Free PMC article. Review. - Molecular basis of RNA recombination in the 3'UTR of chikungunya virus genome.
Bardossy ES, Volpe S, Suzuki Y, Merwaiss F, Faraj S, Montes M, Saleh MC, Alvarez DE, Filomatori CV. Bardossy ES, et al. Nucleic Acids Res. 2024 Sep 9;52(16):9727-9744. doi: 10.1093/nar/gkae650. Nucleic Acids Res. 2024. PMID: 39051569 Free PMC article. - KwARG: parsimonious reconstruction of ancestral recombination graphs with recurrent mutation.
Ignatieva A, Lyngsø RB, Jenkins PA, Hein J. Ignatieva A, et al. Bioinformatics. 2021 Oct 11;37(19):3277-3284. doi: 10.1093/bioinformatics/btab351. Bioinformatics. 2021. PMID: 33970217 Free PMC article. - Detection and prevalence of SARS-CoV-2 co-infections during the Omicron variant circulation in France.
Bal A, Simon B, Destras G, Chalvignac R, Semanas Q, Oblette A, Quéromès G, Fanget R, Regue H, Morfin F, Valette M, Lina B, Josset L. Bal A, et al. Nat Commun. 2022 Oct 23;13(1):6316. doi: 10.1038/s41467-022-33910-9. Nat Commun. 2022. PMID: 36274062 Free PMC article. - Helicase-mediated changes in RNA structure at the single-molecule level.
König SL, Liyanage PS, Sigel RK, Rueda D. König SL, et al. RNA Biol. 2013 Jan;10(1):133-48. doi: 10.4161/rna.23507. Epub 2013 Jan 1. RNA Biol. 2013. PMID: 23353571 Free PMC article. Review.
References
- Holmes EC. The Evolution and Emergence of RNA Viruses. 2009.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources