Laboratory adaptation of Bordetella pertussis is associated with the loss of type three secretion system functionality - PubMed (original) (raw)
Laboratory adaptation of Bordetella pertussis is associated with the loss of type three secretion system functionality
M E Gaillard et al. Infect Immun. 2011 Sep.
Abstract
Although Bordetella pertussis contains and transcribes loci encoding type III secretion system (TTSS) homologues, expression of TTSS-associated proteins has been reported only for non-laboratory-adapted Irish clinical isolates. Here we confirm such a result for clinical isolates obtained from patients treated in Argentinean hospitals. Moreover, we demonstrate that the expression of TTSS-associated proteins is independent both of the year in which the isolate was obtained and of the types of polymorphic alleles for other virulence factors but is dependent on environmental growth conditions. Interestingly, we observed that TTSS-associated protein expression is lost after successive in vitro passages but becomes operative again when bacteria come into contact with the host. This in vivo activation of TTSS expression was observed not only for clinical isolates previously adapted to the laboratory after successive in vitro passages but also for vaccine strains that did not express the system in vitro. The reversibility of TTSS expression, demonstrated by its switching off-on when the bacterium comes into contact with the host, appears to be an adaptive response of this pathogen.
Figures
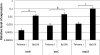
Fig. 1.
Quantitative PCR analysis of bsc N, bsc C, and _bsp_22 transcript levels in the vaccine strain Tohama I and the clinical isolate _Bp_106. cDNA obtained from total RNA of Tohama I and _Bp_106 was used as a template for PCR. The rec A transcript was used as a reference. Comparisons of Tohama I with _Bp_106 were performed for each gene evaluated; P values were <0.05 (a and b) and <0.01 (c).
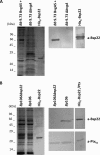
Fig. 2.
Reactivities of an anti-Bsp22 serum against supernatant proteins from B. bronchiseptica and B. pertussis strains. Proteins from B. bronchiseptica 9.73 grown under Bvg+ conditions (_Bb_9.73 BvgAS+) and from its avirulent constitutive derivative mutant (Bb_9.73 Δ_bvgA) (A) and from a Bsp22-defective mutant of B. pertussis Bp106 (Bp_106 Δ_bsp22) and its parental strain _Bp_106 (B) were resolved by 15% (wt/vol) SDS-PAGE (left) and were probed with the indicated sera (right). Purified His6-Bsp22 and Ptx were used as positive controls.
Fig. 3.
Immunoblot analysis of supernatant proteins from Argentinean B. pertussis clinical isolates and laboratory-adapted strains. Proteins from reference and vaccine strains (A) and from clinical isolates (B) were resolved by 15% (wt/vol) SDS-PAGE, transferred to a PVDF membrane, and probed with a polyclonal serum specific for Bsp22. Samples in panel A were also probed with a polyclonal antiserum specific for subunit S1 of pertussis toxin (PtxS1). Positive controls were purified recombinant His6-Bsp22 and PtxS1 proteins, as well as samples obtained from B. bronchiseptica strain 9.73 (_Bb_9.73) and the clinical isolate _Bp_106.
Fig. 4.
Immunoblot analysis of laboratory-adapted B. pertussis isolates/strains for TTSS expression. (A) Supernatant proteins of B. pertussis strains _Bp_6901, _Bp_106, and Tohama I with their derivatives obtained after 35 successive in vitro passages (Adapted). (B) TTSS expression through 35 in vitro passages of _Bp_106. The sera used in immunoblotting are indicated on the left.
Fig. 5.
Immunoblot analysis of TTSS expression through 5 in vitro passages of _Bp_106. Supernatant proteins of B. pertussis Bp106 with their derivatives obtained after 5 successive in vitro passages were resolved by 15% (wt/vol) SDS-PAGE, transferred to a PVDF membrane, and probed with a polyclonal serum specific for Bsp22. Samples were also probed with a polyclonal antiserum specific for subunit S1 of pertussis toxin (PtxS1). Purified recombinant His6-Bsp22 and PtxS1 proteins were included in this assay as positive controls.
Fig. 6.
Analysis of TTSS expression in vivo. Protein samples obtained from B. pertussis supernatant cultures (in vitro) or from whole-cell bacteria recovered from infected mice (in vivo) were analyzed for Bsp22 expression by immunoblotting. Samples obtained from Tohama I, _Bp_106 (_Bp_106 Not Adapted), and isolate _Bp_106 subjected to 35 in vitro passages (_Bp_106 Adapted) were probed with a polyclonal antiserum specific for Bsp22 or subunit S1 of pertussis toxin (PtxS1). Purified His6-Bsp22 and Ptx were used as positive controls.
Similar articles
- Bordetella pertussis expresses a functional type III secretion system that subverts protective innate and adaptive immune responses.
Fennelly NK, Sisti F, Higgins SC, Ross PJ, van der Heide H, Mooi FR, Boyd A, Mills KH. Fennelly NK, et al. Infect Immun. 2008 Mar;76(3):1257-66. doi: 10.1128/IAI.00836-07. Epub 2008 Jan 14. Infect Immun. 2008. PMID: 18195025 Free PMC article. - Transcriptional profiling of Bordetella pertussis reveals requirement of RNA chaperone Hfq for Type III secretion system functionality.
Bibova I, Hot D, Keidel K, Amman F, Slupek S, Cerny O, Gross R, Vecerek B. Bibova I, et al. RNA Biol. 2015;12(2):175-85. doi: 10.1080/15476286.2015.1017237. RNA Biol. 2015. PMID: 25674816 Free PMC article. - Demonstration of differential virulence gene promoter activation in vivo in Bordetella pertussis using RIVET.
Veal-Carr WL, Stibitz S. Veal-Carr WL, et al. Mol Microbiol. 2005 Feb;55(3):788-98. doi: 10.1111/j.1365-2958.2004.04418.x. Mol Microbiol. 2005. PMID: 15661004 - Pertussis pathogenesis--what we know and what we don't know.
Hewlett EL, Burns DL, Cotter PA, Harvill ET, Merkel TJ, Quinn CP, Stibitz ES. Hewlett EL, et al. J Infect Dis. 2014 Apr 1;209(7):982-5. doi: 10.1093/infdis/jit639. J Infect Dis. 2014. PMID: 24626533 Free PMC article. Review. - Bordetella Type III Secretion Injectosome and Effector Proteins.
Kamanova J. Kamanova J. Front Cell Infect Microbiol. 2020 Sep 4;10:466. doi: 10.3389/fcimb.2020.00466. eCollection 2020. Front Cell Infect Microbiol. 2020. PMID: 33014891 Free PMC article. Review.
Cited by
- T3SS chaperone of the CesT family is required for secretion of the anti-sigma factor BtrA in Bordetella pertussis.
Držmíšek J, Petráčková D, Dienstbier A, Čurnová I, Večerek B. Držmíšek J, et al. Emerg Microbes Infect. 2023 Dec;12(2):2272638. doi: 10.1080/22221751.2023.2272638. Epub 2023 Nov 1. Emerg Microbes Infect. 2023. PMID: 37850324 Free PMC article. - Cyclic di-GMP Regulates the Type III Secretion System and Virulence in Bordetella bronchiseptica.
Gutierrez MP, Wong TY, Damron FH, Fernández J, Sisti F. Gutierrez MP, et al. Infect Immun. 2022 Jun 16;90(6):e0010722. doi: 10.1128/iai.00107-22. Epub 2022 May 25. Infect Immun. 2022. PMID: 35612302 Free PMC article. - Molecular Targets and Strategies for Inhibition of the Bacterial Type III Secretion System (T3SS); Inhibitors Directly Binding to T3SS Components.
Hotinger JA, Pendergrass HA, May AE. Hotinger JA, et al. Biomolecules. 2021 Feb 19;11(2):316. doi: 10.3390/biom11020316. Biomolecules. 2021. PMID: 33669653 Free PMC article. Review. - Omics Analysis of Blood-Responsive Regulon in Bordetella pertussis Identifies a Novel Essential T3SS Substrate.
Drzmisek J, Stipl D, Petrackova D, Vecerek B, Dienstbier A. Drzmisek J, et al. Int J Mol Sci. 2021 Jan 13;22(2):736. doi: 10.3390/ijms22020736. Int J Mol Sci. 2021. PMID: 33450976 Free PMC article. - Comparative Omics Analysis of Historic and Recent Isolates of Bordetella pertussis and Effects of Genome Rearrangements on Evolution.
Dienstbier A, Amman F, Petráčková D, Štipl D, Čapek J, Zavadilová J, Fabiánová K, Držmíšek J, Kumar D, Wildung M, Pouchnik D, Večerek B. Dienstbier A, et al. Emerg Infect Dis. 2021 Jan;27(1):57-68. doi: 10.3201/eid2701.191541. Emerg Infect Dis. 2021. PMID: 33350934 Free PMC article.
References
- Bouchez V., et al. 2009. First report and detailed characterization of B. pertussis isolates not expressing pertussis toxin or pertactin. Vaccine 27: 6034–6041 - PubMed
- Bradford M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72: 248–254 - PubMed
- Büttner D., Bonas U. 2002. Port of entry—the type III secretion translocon. Trends Microbiol. 10: 186–192 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources