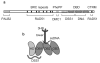Unraveling the mechanism of BRCA2 in homologous recombination - PubMed (original) (raw)
Review
Unraveling the mechanism of BRCA2 in homologous recombination
William K Holloman. Nat Struct Mol Biol. 2011.
Abstract
BRCA2 is the product of a breast cancer susceptibility gene in humans and the founding member of an emerging family of proteins present throughout the eukaryotic domain that serve in homologous recombination. The function of BRCA2 in recombination is to control RAD51, a protein that catalyzes homologous pairing and DNA strand exchange. By physically interacting with both RAD51 and single-stranded DNA, BRCA2 mediates delivery of RAD51 preferentially to sites of single-stranded DNA (ssDNA) exposed as a result of DNA damage or replication problems. Through its action, BRCA2 helps restore and maintain integrity of the genome. This review highlights recent studies on BRCA2 and its orthologs that have begun to illuminate the molecular mechanisms by which these proteins control homologous recombination.
Figures
Figure 1. DNA repair by HR
Schematics are shown illustrating how repair of DNA with a DSB or ssDNA gap could initiate by HR. Both pathways begin with invasion of a ssDNA tract (step i or i-a) into a homologous DNA sequence. In DSB repair the D-loop formed after invasion of the ssDNA tail (step ii) is extended by DNA synthesis (step iii) until it can pair with, or capture, the second end through exposed complementary sequence. The 3′ strand of the second end serves as a primer for fill-in synthesis (step iv). Additional processing of the intermediates and several subsequent steps results in a repaired chromosome. In the other pathway, termed post-replication repair, a ssDNA gap generated during DNA replication of a damaged template (step i-a) invades the undamaged homologous sequence present in the sister chromatid (step ii-a). By branch migration through a short tract the 3′ end of the broken strand switches templates (step iii-a) and then serves as primer for fill-in synthesis (step iv-a). After additional steps the intermediate is resolved and repair is completed.
Figure 2. BRCA2 organization and DBD domain structure
(a) BRCA2 is shown schematically with protein interaction motifs and domains identified on top. The acronyms are as described in the text. CTRM is
C
-
t
erminal
R
AD51-binding
m
otif. The elements are illustrated as gray and black boxes. The DBD is broken down into the helix-rich domain (hatches) and the three OB folds (ovals). The approximate regions of interaction with the various proteins and DNA discussed are shown underneath. (b) The 800 residue DBD is shown schematically with the helix-rich domain (HD) followed by the OB folds. OB2 and OB3 are packed in tandem while OB1 is packed with OB2 in the opposite orientation. The Tower domain emerges from OB2 and has a three-helix bundle (3HB) on top. ssDNA (black squiggle) interacts with OB2 and OB3. DSS1 (gray squiggle) interacts with HD and OB1 on the opposite face of the domain.
Similar articles
- Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA.
Liu J, Doty T, Gibson B, Heyer WD. Liu J, et al. Nat Struct Mol Biol. 2010 Oct;17(10):1260-2. doi: 10.1038/nsmb.1904. Epub 2010 Aug 22. Nat Struct Mol Biol. 2010. PMID: 20729859 Free PMC article. - Structure and mechanism of action of the BRCA2 breast cancer tumor suppressor.
Shahid T, Soroka J, Kong E, Malivert L, McIlwraith MJ, Pape T, West SC, Zhang X. Shahid T, et al. Nat Struct Mol Biol. 2014 Nov;21(11):962-968. doi: 10.1038/nsmb.2899. Epub 2014 Oct 5. Nat Struct Mol Biol. 2014. PMID: 25282148 Free PMC article. - The breast cancer tumor suppressor BRCA2 promotes the specific targeting of RAD51 to single-stranded DNA.
Thorslund T, McIlwraith MJ, Compton SA, Lekomtsev S, Petronczki M, Griffith JD, West SC. Thorslund T, et al. Nat Struct Mol Biol. 2010 Oct;17(10):1263-5. doi: 10.1038/nsmb.1905. Epub 2010 Aug 22. Nat Struct Mol Biol. 2010. PMID: 20729858 Free PMC article. - Structural insights into BRCA2 function.
Shamoo Y. Shamoo Y. Curr Opin Struct Biol. 2003 Apr;13(2):206-11. doi: 10.1016/s0959-440x(03)00033-2. Curr Opin Struct Biol. 2003. PMID: 12727514 Review. - RecA: Regulation and Mechanism of a Molecular Search Engine.
Bell JC, Kowalczykowski SC. Bell JC, et al. Trends Biochem Sci. 2016 Jun;41(6):491-507. doi: 10.1016/j.tibs.2016.04.002. Epub 2016 May 4. Trends Biochem Sci. 2016. PMID: 27156117 Free PMC article. Review.
Cited by
- DSS1 restrains BRCA2's engagement with dsDNA for homologous recombination, replication fork protection, and R-loop homeostasis.
Huang Y, Li W, Foo T, Ji JH, Wu B, Tomimatsu N, Fang Q, Gao B, Long M, Xu J, Maqbool R, Mukherjee B, Ni T, Alejo S, He Y, Burma S, Lan L, Xia B, Zhao W. Huang Y, et al. Nat Commun. 2024 Aug 17;15(1):7081. doi: 10.1038/s41467-024-51557-6. Nat Commun. 2024. PMID: 39152168 Free PMC article. - Predictive modeling of gene mutations for the survival outcomes of epithelial ovarian cancer patients.
Ma MC, Lavi ES, Altwerger G, Lin ZP, Ratner ES. Ma MC, et al. PLoS One. 2024 Jul 8;19(7):e0305273. doi: 10.1371/journal.pone.0305273. eCollection 2024. PLoS One. 2024. PMID: 38976671 Free PMC article. - Therapeutic Targeting of DNA Repair Pathways in Pediatric Extracranial Solid Tumors: Current State and Implications for Immunotherapy.
Zhao SJ, Prior D, Heske CM, Vasquez JC. Zhao SJ, et al. Cancers (Basel). 2024 Apr 25;16(9):1648. doi: 10.3390/cancers16091648. Cancers (Basel). 2024. PMID: 38730598 Free PMC article. Review. - Genomic Features of Homologous Recombination Deficiency in Breast Cancer: Impact on Testing and Immunotherapy.
Ali U, Vungarala S, Tiriveedhi V. Ali U, et al. Genes (Basel). 2024 Jan 26;15(2):162. doi: 10.3390/genes15020162. Genes (Basel). 2024. PMID: 38397152 Free PMC article. Review. - Case report of penile squamous cell carcinoma continuous treatment with BRCA2 mutation.
Zhang Q, Li Y, Zhang Y, Deng Z, Ding Y. Zhang Q, et al. World J Surg Oncol. 2024 Feb 9;22(1):50. doi: 10.1186/s12957-024-03305-9. World J Surg Oncol. 2024. PMID: 38336701 Free PMC article.
References
- Budzowska M, Kanaar R. Mechanisms of dealing with DNA damage-induced replication problems. Cell Biochem Biophys. 2009;53:17–31. - PubMed
- Mimitou EP, Symington LS. Nucleases and helicases take center stage in homologous recombination. Trends Biochem Sci. 2009;34:264–72. - PubMed
- San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem. 2008;77:229–57. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous