Role of autophagy in cancer prevention - PubMed (original) (raw)
Review
Role of autophagy in cancer prevention
Hsin-Yi Chen et al. Cancer Prev Res (Phila). 2011 Jul.
Abstract
Macroautophagy (autophagy hereafter) is a catabolic process by which cells degrade intracellular components in lysosomes. This cellular garbage disposal and intracellular recycling system maintains cellular homeostasis by eliminating superfluous or damaged proteins and organelles and invading microbes and by providing substrates for energy generation and biosynthesis in stress. Autophagy thus promotes the health of cells and animals and is critical for the development, differentiation, and maintenance of cell function and for the host defense against pathogens. Deregulation of autophagy is linked to susceptibility to various disorders including degenerative diseases, metabolic syndrome, aging, infectious diseases, and cancer. Autophagic activity emerges as a critical factor in the development and progression of diseases that are associated with increased cancer risk as well as in different stages of cancer. Given that cancer is a complex process and autophagy exerts its effects in multiple ways, the role of autophagy in tumorigenesis is context-dependent. As a cytoprotective survival pathway, autophagy prevents chronic tissue damage that can lead to cancer initiation and progression. In this setting, stimulation or restoration of autophagy may prevent cancer. In contrast, once cancer occurs, many cancer cells upregulate basal autophagy and utilize autophagy to enhance fitness and survive in the hostile tumor microenvironment. These findings revealed the concept that aggressive cancers can be addicted to autophagy for survival. In this setting, autophagy inhibition is a therapeutic strategy for established cancers.
Figures
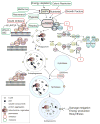
Figure 1. Machinery and small molecule modulators of autophagy
The events of autophagosome formation- nucleation, expansion and maturation are depicted along with molecular machinery that regulates this process. The major negative regulator of autophagy, mTORC1, which integrates stimuli including availability of nutrients or growth factors, energy depletion or hypoxia, is also shown. Drugable protein targets are highlighted with striated outlines. Green or Red boxes indicate autophagy stimulators or inhibitors respectively. Question marks represent inhibitors aiming at potential targets, the kinase Ulk1, the cysteine protease Atg4 and the E1-like ubiquitination enzyme Atg7. AMPK: AMP-activated protein kinase. PE: phosphatidylethanolamine. IP3: inositol-1,4,5-triphosphate. IP3R: inositol-1,4,5-triphosphate receptor. CBZ: carbamazepine. HCQ: hydroxychloroquine.
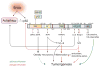
Figure 2. Role of autophagic regulation of p62 in cell signaling
Stress upregulates autophagy and also induces the accumulation of the autophagic cargo receptor protein p62. Autophagy regulates p62 turnover and its protein levels in cells (denoted by an asterisk). Domains of p62 are depicted diagrammatically. PB1: Phox and Bem1p-1 oligomerization domain. ZZ: zinc finger. TBS: TRAF6-binding sequence. LIR: LC3-interacting region. KIR: Keap1-interacting region. UBA: ubiquitin-associated domain. Numbers represent the corresponding amino acid positions in mouse p62. NLS: nuclear localization signal. NES: nuclear export signal (40, 43, 76). Brown arrows indicate p62-protein interactions. The dash indicates a putative interaction between p62 and ERK (77). Accumulation of p62 activates Nrf2, while p62 itself is a transcriptional target of Nrf2, creating a positive feedback loop (45). Green or red depicts signals occurring when p62 loses or gains function, respectively. The dashed green line shows a putative role of obesity in tumorigenesis (see Fig. 3). The dashed red line shows a putative consequence of hyperactive Nrf2.
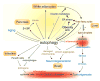
Figure 3. Role of autophagy in health and disease progression
Autophagy is required for cellular and organismal heath and is involved in the pathogenesis of a variety of diseases. Disorders are shown in blue. Lines with an arrowhead or a cross short line represent positive or negative regulation of the pathway, respectively. Bold or gray lines represent pathways upregulated or downregulated respectively during lipid homeostasis collapse. The dashed line depicts flux of fatty acids from adipocytes to liver. The asterisk indicates the abnormal deposit of fatty acids in liver when lipid homeostasis is disrupted. The question mark denotes a putative regulation. Yellow shading shows where autophagy is involved in the regulation of lipid homeostasis. Red shading shows inflammation, which promotes tumorigenesis and is suppressed by autophagy. HFD: high-fat diet.
Similar articles
- Autophagy, Metabolism, and Cancer.
White E, Mehnert JM, Chan CS. White E, et al. Clin Cancer Res. 2015 Nov 15;21(22):5037-46. doi: 10.1158/1078-0432.CCR-15-0490. Clin Cancer Res. 2015. PMID: 26567363 Free PMC article. Review. - Autophagy and cancer cell metabolism.
Lozy F, Karantza V. Lozy F, et al. Semin Cell Dev Biol. 2012 Jun;23(4):395-401. doi: 10.1016/j.semcdb.2012.01.005. Epub 2012 Jan 18. Semin Cell Dev Biol. 2012. PMID: 22281437 Free PMC article. Review. - The role for autophagy in cancer.
White E. White E. J Clin Invest. 2015 Jan;125(1):42-6. doi: 10.1172/JCI73941. Epub 2015 Jan 2. J Clin Invest. 2015. PMID: 25654549 Free PMC article. Review. - Xenophagy in cancer.
Ammanathan V, Vats S, Abraham IM, Manjithaya R. Ammanathan V, et al. Semin Cancer Biol. 2020 Nov;66:163-170. doi: 10.1016/j.semcancer.2020.02.015. Epub 2020 Feb 29. Semin Cancer Biol. 2020. PMID: 32126260 Review. - Deconvoluting the context-dependent role for autophagy in cancer.
White E. White E. Nat Rev Cancer. 2012 Apr 26;12(6):401-10. doi: 10.1038/nrc3262. Nat Rev Cancer. 2012. PMID: 22534666 Free PMC article. Review.
Cited by
- COX7A1 suppresses the viability of human non-small cell lung cancer cells via regulating autophagy.
Zhao L, Chen X, Feng Y, Wang G, Nawaz I, Hu L, Liu P. Zhao L, et al. Cancer Med. 2019 Dec;8(18):7762-7773. doi: 10.1002/cam4.2659. Epub 2019 Oct 30. Cancer Med. 2019. PMID: 31663688 Free PMC article. - CNOT2 promotes degradation of p62/SQSTM1 as a negative regulator in ATG5 dependent autophagy.
Jeong K, Kwon HY, Jeong MS, Sohn EJ, Kim SH. Jeong K, et al. Oncotarget. 2017 Jul 11;8(28):46034-46046. doi: 10.18632/oncotarget.17682. Oncotarget. 2017. PMID: 28537904 Free PMC article. - CaMKII-mediated Beclin 1 phosphorylation regulates autophagy that promotes degradation of Id and neuroblastoma cell differentiation.
Li X, Wu XQ, Deng R, Li DD, Tang J, Chen WD, Chen JH, Ji J, Jiao L, Jiang S, Yang F, Feng GK, Senthilkumar R, Yue F, Zhang HL, Wu RY, Yu Y, Xu XL, Mai J, Li ZL, Peng XD, Huang Y, Huang X, Ma NF, Tao Q, Zeng YX, Zhu XF. Li X, et al. Nat Commun. 2017 Oct 27;8(1):1159. doi: 10.1038/s41467-017-01272-2. Nat Commun. 2017. PMID: 29079782 Free PMC article. - Overaccumulation of p53-mediated autophagy protects against betulinic acid-induced apoptotic cell death in colorectal cancer cells.
Wang S, Wang K, Zhang C, Zhang W, Xu Q, Wang Y, Zhang Y, Li Y, Zhang Y, Zhu H, Song F, Lei Y, Bu Y. Wang S, et al. Cell Death Dis. 2017 Oct 5;8(10):e3087. doi: 10.1038/cddis.2017.485. Cell Death Dis. 2017. PMID: 28981110 Free PMC article. - Sphingolipids as cell fate regulators in lung development and disease.
Lee J, Yeganeh B, Ermini L, Post M. Lee J, et al. Apoptosis. 2015 May;20(5):740-57. doi: 10.1007/s10495-015-1112-6. Apoptosis. 2015. PMID: 25753687 Free PMC article. Review.
References
- Li Y, Wang Y, Kim E, Beemiller P, Wang CY, Swanson J, et al. Bnip3 mediates the hypoxia-induced inhibition on mammalian target of rapamycin by interacting with Rheb. J Biol Chem. 2007;282:35803–13. - PubMed
- Mazure NM, Pouyssegur J. Hypoxia-induced autophagy: cell death or cell survival? Curr Opin Cell Biol. 2010;22:177–80. - PubMed
Publication types
MeSH terms
Grants and funding
- RC1 CA147961/CA/NCI NIH HHS/United States
- R37 CA53370/CA/NCI NIH HHS/United States
- RC1 CA147961-02/CA/NCI NIH HHS/United States
- R01 CA130893-02S1/CA/NCI NIH HHS/United States
- R01 CA130893/CA/NCI NIH HHS/United States
- R01 CA130893-03/CA/NCI NIH HHS/United States
- R37 CA053370/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials