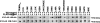Contribution of E3-ubiquitin ligase activity to HIV-1 restriction by TRIM5alpha(rh): structure of the RING domain of TRIM5alpha - PubMed (original) (raw)
Contribution of E3-ubiquitin ligase activity to HIV-1 restriction by TRIM5alpha(rh): structure of the RING domain of TRIM5alpha
Maritza Lienlaf et al. J Virol. 2011 Sep.
Abstract
TRIM5α(rh) is a cytosolic protein that potently restricts HIV-1 before reverse transcription. TRIM5α(rh) is composed of four different domains: RING, B-box 2, coiled coil, and B30.2(SPRY). The contribution of each of these domains to restriction has been extensively studied, with the exception of the RING domain. The RING domain of TRIM5α exhibits E3-ubiquitin ligase activity, but the contribution of this activity to the restriction of HIV-1 is not known. To test the hypothesis that the E3-ubiquitin ligase activity of the RING domain modulates TRIM5α(rh) restriction of HIV-1, we correlated the E3-ubiquitin ligase activity of a panel of TRIM5α(rh) RING domain variants with the ability of these mutant proteins to restrict HIV-1. For this purpose, we first solved the nuclear magnetic resonance structure of the RING domain of TRIM5α and defined potential functional regions of the RING domain by homology to other RING domains. With this structural information, we performed a systematic mutagenesis of the RING domain regions and tested the TRIM5α RING domain variants for the ability to undergo self-ubiquitylation. Several residues, particularly the ones on the E2-binding region of the RING domain, were defective in their self-ubiquitylation ability. To correlate HIV-1 restriction to self-ubiquitylation, we used RING domain mutant proteins that were defective in self-ubiquitylation but preserve important properties required for potent restriction by TRIM5α(rh), such as capsid binding and higher-order self-association. From these investigations, we found a set of residues that when mutated results in TRIM5α molecules that lost both the ability to potently restrict HIV-1 and their self-ubiquitylation activity. Remarkably, all of these changes were in residues located in the E2-binding region of the RING domain. Overall, these results demonstrate a role for TRIM5α self-ubiquitylation in the ability of TRIM5α to restrict HIV-1.
Figures
Fig. 1.
Solution structure of the human TRIM5α RING domain. (A) Superposition of 20 NMR structures showing the α-helix in red, β-strands in blue, and Zn2+ in green. (B) Ribbon diagram of the NMR structure shown in panel A from the same perspective. The Zn2+ coordinating residues are also shown in green. (C) Electrostatic mapping of the RING domain surface highlighting the positions of positive (blue), negative (red), and neutral (white) charges. Dotted circles indicate the location of the putative E2-binding site for the RING domain of TRIM5α. (D) A Corey-Pauling-Koltun model of the RING domain of human TRIM5α is shown with labels of the visualized residues. Acidic and basic residues are shown in red and blue, respectively. Hydrophobic residues are shown in green.
Fig. 2.
The putative E2-binding and RING-RING interaction regions of the TRIM5αrh RING domain. The structure of the TRIM5αrh RING domain is based on the human TRIM5α RING domain (PDB 2ECV) and was assembled by using the SWISS-MODEL protein homology modeling program. (A) The putative E2-binding region (magenta) was identified by fitting the model structure to the Cbl-UBCH7 complex structure and by modeling the interaction between the RING domain and UBCH7. (B) The putative RING-RING interaction region (green) was identified in the same way by fitting it to the BRCA1-BARD1 RING structure for the RING domain region. Since conformational prediction of the N-terminal and C-terminal regions was difficult due to poor sequence homology, these regions were not included. Residues that interact with Zn2+ atoms are indicated by black asterisks above the sequence alignment.
Fig. 3.
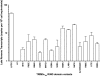
E3-ubiquitin ligase activities of TRIM5αrh RING domain variants. Human 293T cells were transfected with plasmids encoding FLAG-tagged mutant and wild-type TRIM5αrh proteins. Forty-eight hours later, the cells expressing each TRIM5αrh variant were lysed in whole-cell extract and immunoprecipitated using anti-FLAG–agarose beads as described in Materials and Methods. Beads containing the immunoprecipitated TRIM5αrh variants were washed and eluted using 200 μg/ml FLAG tripeptide in whole-cell extract buffer as described in Materials and Methods. Samples were supplemented with 5 μM ubiquitin aldehyde, a potent inhibitor of all ubiquitin C-terminal hydrolases, ubiquitin-specific proteases, and deubiquitinating enzymes. Similar amounts of inhibitor-treated samples containing mutant and wild-type TRIM5αrh were incubated with or without 200 nM enzyme E1 (human recombinant UBE1) and 100 nM enzyme E2 (human recombinant UbcH5a) as indicated. Reaction mixtures were supplemented with 200 μM ubiquitin tagged with a myc epitope (human recombinant ubiquitin) and an energy regeneration solution containing MgCl2, ATP, and ATP-regenerating enzymes to recycle hydrolyzed ATP. The reaction mixture was incubated at 37°C for 1 h, and collected fractions were analyzed by Western blotting using HRP-conjugated antibodies against FLAG to detect the levels of TRIM5αrh variants. To detect ubiquitylated forms of TRIM5αrh variants, membranes were blotted using HRP-conjugated antibodies against myc. Purple circles and green circles indicate TRIM5αrh variants with defective self-ubiquitylation activity located on the E2-binding and RING-RING interaction region, respectively. The results of three independent experiments were similar; the result of a single experiment is shown.
Fig. 4.
Binding of TRIM5αrh RING mutant proteins to assembled HIV-1 capsids. 293T cells were transfected with plasmids expressing the indicated wild-type and mutant TRIM5αrh proteins tagged with HA epitopes. Thirty-six hours after transfection, cells were lysed. The lysates were incubated at room temperature for 1 h with HIV-1 CA-NC complexes that had been assembled in vitro. The mixtures were applied to a 70% sucrose cushion and centrifuged. INPUT represents the lysates analyzed by Western blotting (WB) before being applied to the 70% cushion. The input mixtures were Western blotted for the HA tag. The pellet from the 70% cushion (PELLET) was analyzed by Western blotting using antibodies against the HA tag and HIV-1 CA-NC protein. The Western blots were quantitated as described in Materials and Methods, and binding values are shown in Table 2. The results of three independent experiments were similar; the result of a single experiment is shown.
Fig. 5.
Higher-order self-association of TRIM5αrh RING mutant proteins. 293T cells were transfected with plasmids expressing the indicated wild-type or mutant TRIM5α proteins with a FLAG or an HA epitope tag. Cells expressing wild-type and mutant TRIM5αrh proteins were lysed 48 h after transfection. The cell lysates containing similar inputs were mixed, and the indicated mixtures were used for immunoprecipitation (IP) with an antibody directed against the FLAG epitope, as described in Materials and Methods. Elution of the immunocomplexes was performed with a FLAG tripeptide and analyzed by Western blotting (WB) using anti-HA and anti-FLAG antibodies. The results of three independent experiments were similar; the result of a single experiment is shown.
Fig. 6.
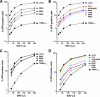
Restriction of HIV-1 and EIAV infection by TRIM5αrh mutant proteins. Cf2Th cells were transduced with the LPCX vector expressing HA-tagged wild-type and mutant TRIM5αrh proteins. Stable cell lines were selected with 5 μg/ml puromycin, and the expression levels of mutant and wild-type TRIM5αrh proteins were assayed by Western blotting using HRP-conjugated antibodies against HA (see Fig. S4 in the supplemental material). The cells were challenged with different amounts of HIV-1–GFP (A, B) or EIAV-GFP (C, D). The percentage of GFP-positive cells was measured 48 h later by FACS. The results of three independent experiments were similar; the results of a single experiment are shown.
Fig. 7.
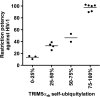
TRIM5α self-ubiquitylation activity correlates with anti-HIV-1 activity. The abilities of RING domain variants to self-ubiquitylate (TRIM5α self-ubiquitylation) and restrict HIV-1 were assessed as described in the footnotes to Table 2 and in Materials and Methods. TRIM5α RING domain variants that were not defective in binding to the HIV-1 capsid and higher-order self-association were analyzed. The Spearman rank correlation coefficient, rs, is 0.9090, with a 95% confidence interval of 0.8623 to 0.9847 (two-sided P value of < 0.0001).
Fig. 8.
Blockade of HIV-1 reverse transcription by TRIM5αrh RING mutant proteins. Cf2Th cells expressing the indicated wild-type and mutant TRIM5αrh proteins or containing the empty LPCX vector were challenged at an MOI of 0.4 with DNase-pretreated HIV-1–GFP. After 7 h, cells were lysed and total DNA was extracted. The levels of viral DNA were measured by quantitative real-time PCR using a probe against GFP as described in Materials and Methods. Similar results were obtained in three independent experiments.
Similar articles
- Virus-specific effects of TRIM5α(rh) RING domain functions on restriction of retroviruses.
Li X, Kim J, Song B, Finzi A, Pacheco B, Sodroski J. Li X, et al. J Virol. 2013 Jul;87(13):7234-45. doi: 10.1128/JVI.00620-13. Epub 2013 May 1. J Virol. 2013. PMID: 23637418 Free PMC article. - Role of TRIM5α RING domain E3 ubiquitin ligase activity in capsid disassembly, reverse transcription blockade, and restriction of simian immunodeficiency virus.
Kim J, Tipper C, Sodroski J. Kim J, et al. J Virol. 2011 Aug;85(16):8116-32. doi: 10.1128/JVI.00341-11. Epub 2011 Jun 15. J Virol. 2011. PMID: 21680520 Free PMC article. - A B-box 2 surface patch important for TRIM5alpha self-association, capsid binding avidity, and retrovirus restriction.
Diaz-Griffero F, Qin XR, Hayashi F, Kigawa T, Finzi A, Sarnak Z, Lienlaf M, Yokoyama S, Sodroski J. Diaz-Griffero F, et al. J Virol. 2009 Oct;83(20):10737-51. doi: 10.1128/JVI.01307-09. Epub 2009 Aug 5. J Virol. 2009. PMID: 19656869 Free PMC article. - TRIM5alpha.
Song B. Song B. Curr Top Microbiol Immunol. 2009;339:47-66. doi: 10.1007/978-3-642-02175-6_3. Curr Top Microbiol Immunol. 2009. PMID: 20012523 Review. - Anti-retroviral activity of TRIM5 alpha.
Nakayama EE, Shioda T. Nakayama EE, et al. Rev Med Virol. 2010 Mar;20(2):77-92. doi: 10.1002/rmv.637. Rev Med Virol. 2010. PMID: 20049904 Review.
Cited by
- Contribution of SUMO-interacting motifs and SUMOylation to the antiretroviral properties of TRIM5α.
Brandariz-Nuñez A, Roa A, Valle-Casuso JC, Biris N, Ivanov D, Diaz-Griffero F. Brandariz-Nuñez A, et al. Virology. 2013 Jan 20;435(2):463-71. doi: 10.1016/j.virol.2012.09.042. Epub 2012 Oct 16. Virology. 2013. PMID: 23084420 Free PMC article. - The V86M mutation in HIV-1 capsid confers resistance to TRIM5α by abrogation of cyclophilin A-dependent restriction and enhancement of viral nuclear import.
Veillette M, Bichel K, Pawlica P, Freund SM, Plourde MB, Pham QT, Reyes-Moreno C, James LC, Berthoux L. Veillette M, et al. Retrovirology. 2013 Feb 28;10:25. doi: 10.1186/1742-4690-10-25. Retrovirology. 2013. PMID: 23448277 Free PMC article. - Structural and functional asymmetry of RING trimerization controls priming and extension events in TRIM5α autoubiquitylation.
Herkules F, Yu CH, Taylor AB, Dougherty V, Weintraub ST, Ivanov DN. Herkules F, et al. Nat Commun. 2022 Nov 19;13(1):7104. doi: 10.1038/s41467-022-34920-3. Nat Commun. 2022. PMID: 36402777 Free PMC article. - Using antiubiquitin antibodies to probe the ubiquitination state within rhTRIM5α cytoplasmic bodies.
Danielson CM, Hope TJ. Danielson CM, et al. AIDS Res Hum Retroviruses. 2013 Oct;29(10):1373-85. doi: 10.1089/AID.2013.0029. Epub 2013 Jul 26. AIDS Res Hum Retroviruses. 2013. PMID: 23799296 Free PMC article. - Human nucleoporins promote HIV-1 docking at the nuclear pore, nuclear import and integration.
Di Nunzio F, Danckaert A, Fricke T, Perez P, Fernandez J, Perret E, Roux P, Shorte S, Charneau P, Diaz-Griffero F, Arhel NJ. Di Nunzio F, et al. PLoS One. 2012;7(9):e46037. doi: 10.1371/journal.pone.0046037. Epub 2012 Sep 25. PLoS One. 2012. PMID: 23049930 Free PMC article.
References
- Bellon S. F., Rodgers K. K., Schatz D. G., Coleman J. E., Steitz T. A. 1997. Crystal structure of the RAG1 dimerization domain reveals multiple zinc-binding motifs including a novel zinc binuclear cluster. Nat. Struct. Biol. 4:586–591 - PubMed
- Best S., Le Tissier P., Towers G., Stoye J. P. 1996. Positional cloning of the mouse retrovirus restriction gene Fv1. Nature 382:826–829 - PubMed
- Borden K. L. 2000. RING domains: master builders of molecular scaffolds? J. Mol. Biol. 295:1103–1112 - PubMed
- Borden K. L. 1998. RING fingers and B-boxes: zinc-binding protein-protein interaction domains. Biochem. Cell Biol. 76:351–358 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R00 MH086162/MH/NIMH NIH HHS/United States
- R01 AI087390/AI/NIAID NIH HHS/United States
- 4R00MH086162-02/MH/NIMH NIH HHS/United States
- R01AI7930231/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases