Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis - PubMed (original) (raw)
. 2011 Jul 6;475(7354):106-9.
doi: 10.1038/nature10189.
Florian A Karreth, Timothy J Humpton, Aarthi Gopinathan, Cong Wei, Kristopher Frese, Dipti Mangal, Kenneth H Yu, Charles J Yeo, Eric S Calhoun, Francesca Scrimieri, Jordan M Winter, Ralph H Hruban, Christine Iacobuzio-Donahue, Scott E Kern, Ian A Blair, David A Tuveson
Affiliations
- PMID: 21734707
- PMCID: PMC3404470
- DOI: 10.1038/nature10189
Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis
Gina M DeNicola et al. Nature. 2011.
Abstract
Reactive oxygen species (ROS) are mutagenic and may thereby promote cancer. Normally, ROS levels are tightly controlled by an inducible antioxidant program that responds to cellular stressors and is predominantly regulated by the transcription factor Nrf2 (also known as Nfe2l2) and its repressor protein Keap1 (refs 2-5). In contrast to the acute physiological regulation of Nrf2, in neoplasia there is evidence for increased basal activation of Nrf2. Indeed, somatic mutations that disrupt the Nrf2-Keap1 interaction to stabilize Nrf2 and increase the constitutive transcription of Nrf2 target genes were recently identified, indicating that enhanced ROS detoxification and additional Nrf2 functions may in fact be pro-tumorigenic. Here, we investigated ROS metabolism in primary murine cells following the expression of endogenous oncogenic alleles of Kras, Braf and Myc, and found that ROS are actively suppressed by these oncogenes. K-Ras(G12D), B-Raf(V619E) and Myc(ERT2) each increased the transcription of Nrf2 to stably elevate the basal Nrf2 antioxidant program and thereby lower intracellular ROS and confer a more reduced intracellular environment. Oncogene-directed increased expression of Nrf2 is a new mechanism for the activation of the Nrf2 antioxidant program, and is evident in primary cells and tissues of mice expressing K-Ras(G12D) and B-Raf(V619E), and in human pancreatic cancer. Furthermore, genetic targeting of the Nrf2 pathway impairs K-Ras(G12D)-induced proliferation and tumorigenesis in vivo. Thus, the Nrf2 antioxidant and cellular detoxification program represents a previously unappreciated mediator of oncogenesis.
©2011 Macmillan Publishers Limited. All rights reserved
Figures
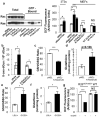
Figure 1. Physiological expression of oncogenes lowers ROS
NIH3T3s and MEFs were transduced with retroviral vectors and evaluated 6 days later: control vector (pBabe), pBabe-H-RasG12V (p-Babe-H-Ras), or pBabe-K-RasG12D (p-Babe-K-Ras). Alternatively, LSL-K-RasG12D MEFs were infected with Ad-mock (K-RasLSL/+) or Ad-cre (K-RasG12D/+) and evaluated 4 days later. Wild-type MEFs were infected with Ad-mock (WT Ad-Mock) or Ad-cre (WT Ad-Cre) and used as controls. a, (Left) Western blot of total and GTP-bound Ras in MEFs expressing endogenous and ectopic Ras, with Rac used as a loading control. (Right) ROS levels following expression of oncogenic Ras, as determined by 2′,7′-dichlorofluorescein diacetate (DCF) staining. b, 8-oxo-dGuo levels following ectopic and endogenous expression of K-RasG12D. c-f, Determination of the GSH/GSSG ratios and total cellular glutathione in cells overexpressing ectopic K-RasG12D (c,d), or expressing endogenous K-RasG12D (e,f). g, ROS levels following activation of MycERT2. R26MER/MER MEFs were treated with DMSO or 100nM 4-OHT and assayed after 24 hours. Data are representative of 3 or more independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001 and error bars represent ± SEM here and for all figures.
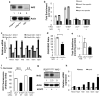
Figure 2. Physiological expression of oncogenes activates the Nrf2 antioxidant program
a, Western blot demonstrates a 60% increase in Nrf2 protein following expression of endogenous K-RasG12D. Antibody specificity was confirmed using Nrf2-/- MEFs. b, Nrf2 ChIP followed by q-PCR for the Hmox1 and Nqo1 promoters. Control non-specific primers amplified regions of DNA located 50Kb from the Hmox1 and Nqo1 promoters. c, Expression of Nrf2 and Nrf2 target genes Nqo1, Hmox1, Gclm, Gclc and Ggt1 upon K-RasG12D expression in Nrf2+/+ and Nrf2-/- MEFs. Nrf2 mRNA is relatively unstable but still detectable at low levels in Nrf2-/- MEFs. d-e, Determination of the GSH/GSSG ratio (d) and total glutathione (e) upon K-RasG12D expression in Nrf2-/- MEFs. f, ROS levels following Nrf2 depletion with siRNA. LSL-K-RasG12D MEFs were transfected with non-targeting (NT) or Nrf2 siRNA, infected with Ad-mock or Ad-cre and assayed after 48 hours for DCF oxidation. g, Western blot of Nrf2 protein levels following induction of MycERT2 by 4-OHT. Densitometry shows a 2.3-fold increase. h, Analysis of Nrf2 antioxidant program gene expression following activation of MycERT2. R26MER/MER MEFs were treated with DMSO or 100nM 4-OHT for 24 hours and assayed for antioxidant gene expression. Data is representative of 3 independent experiments.
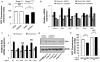
Figure 3. Activation of Nrf2 by K-RasG12D occurs via the Raf-MEK-ERK-Jun pathway
a, ROS levels following treatment of K-RasG12D/+ MEFs with AZD6244. LSL-K-RasG12D MEFs were treated with DMSO or 0.1uM AZD6244, infected with Ad-mock (K-RasLSL/+) or Ad-cre (K-RasG12D/+) and assayed after 72 hours. b, Analysis of antioxidant gene expression following treatment of K-RasLSL/+ and K-RasG12D/+ MEFs with AZD6244 for 24 hours. c, Control of Nrf2 transcription by AP-1 family members. K-RasLSL/+ and K-RasG12D/+ MEFs were transfected with siRNA and assayed for Nrf2 expression after 48 hours. d, Western blot of Jun and actin protein levels in LSL-K-RasG12D and LSL-B-RafV619E MEFs. K-RasLSL/+ and K-RasG12D/+ MEFs were treated with DMSO or 0.1uM AZD6244 for 24 hours. e, ROS levels following Jun depletion with siRNA. LSL-K-RasG12D MEFs were transfected with non-targeting (NT) or Jun siRNA, infected with Ad-mock or Ad-cre and assayed after 48 hours for DCF oxidation. Data are representative of 3 independent experiments.
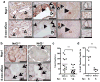
Figure 4. Evidence for Nrf2 antioxidant program in pancreatic cancer
a, Immunohistochemical detection of Nqo1 (brown staining) and 8-oxo-dGuo (purple staining) in mouse PanIN and PDA (similar patterns observed for 11/11 of cases examined) in comparison to morphologically normal ducts. PanIN (arrows), PDA (black arrowheads), normal ducts (white arrowheads) here and for all figures. Scale bar = 56 μm. b, Immunohistochemical detection of Nqo1 and 8-oxo-dGuo in Nrf2-/- PanIN compared to Nrf2+/+ PanIN (similar patterns observed for 5/5 of each genotype examined, PanIN outlined by white dashes). Scale bar = 56 μm. c, Nrf2-/- and Nrf2+/+ PanIN-1a incidence. Whole pancreata were sectioned at 100-micron intervals and total numbers of PanIN-1a were counted. d, Proliferation of PanIN-1a cells in Nrf2-/- and Nrf2+/+ mice, as determined by Ki-67 immunostaining.
Comment in
- Cancer: when antioxidants are bad.
Perera RM, Bardeesy N. Perera RM, et al. Nature. 2011 Jul 6;475(7354):43-4. doi: 10.1038/475043a. Nature. 2011. PMID: 21734699 No abstract available. - Tumorigenesis: oncogene detox programme.
McCarthy N. McCarthy N. Nat Rev Cancer. 2011 Jul 22;11(9):622-3. doi: 10.1038/nrc3119. Nat Rev Cancer. 2011. PMID: 21779002 No abstract available. - NrF2 ⁄Keap1 as gatekeepers of redox homeostasis – do they prevent or cause cancer?
Tew KD. Tew KD. Pigment Cell Melanoma Res. 2011 Dec;24(6):1078-9. doi: 10.1111/j.1755-148X.2011.00913.x. Pigment Cell Melanoma Res. 2011. PMID: 22216435 No abstract available.
Similar articles
- Cancer: when antioxidants are bad.
Perera RM, Bardeesy N. Perera RM, et al. Nature. 2011 Jul 6;475(7354):43-4. doi: 10.1038/475043a. Nature. 2011. PMID: 21734699 No abstract available. - MDA-7/IL-24 inhibits Nrf2-mediated antioxidant response through activation of p38 pathway and inhibition of ERK pathway involved in cancer cell apoptosis.
Tian H, Zhang D, Gao Z, Li H, Zhang B, Zhang Q, Li L, Cheng Q, Pei D, Zheng J. Tian H, et al. Cancer Gene Ther. 2014 Oct;21(10):416-26. doi: 10.1038/cgt.2014.45. Epub 2014 Sep 19. Cancer Gene Ther. 2014. PMID: 25236495 - Impact of Nrf2 on tumour growth and drug sensitivity in oncogenic K-ras-transformed cells in vitro and in vivo.
Shao J, Glorieux C, Liao J, Chen P, Lu W, Liang Z, Wen S, Hu Y, Huang P. Shao J, et al. Free Radic Res. 2018 Jun;52(6):661-671. doi: 10.1080/10715762.2018.1462494. Epub 2018 May 3. Free Radic Res. 2018. PMID: 29621903 - Keap1-Nrf2 signalling in pancreatic cancer.
Hayes AJ, Skouras C, Haugk B, Charnley RM. Hayes AJ, et al. Int J Biochem Cell Biol. 2015 Aug;65:288-99. doi: 10.1016/j.biocel.2015.06.017. Epub 2015 Jun 24. Int J Biochem Cell Biol. 2015. PMID: 26117456 Review. - The Nrf2-antioxidant response element signaling pathway and its activation by oxidative stress.
Nguyen T, Nioi P, Pickett CB. Nguyen T, et al. J Biol Chem. 2009 May 15;284(20):13291-5. doi: 10.1074/jbc.R900010200. Epub 2009 Jan 30. J Biol Chem. 2009. PMID: 19182219 Free PMC article. Review.
Cited by
- Chemical composition of anti-microbially active fractions derived from extract of filamentous fungus Keratinophyton Lemmensii including three novel bioactive compounds.
Labuda R, Bacher M, Rosenau T, Gratzl H, Doppler M, Hager S, Marko D, Wiesner C, Očková M, Ollinger N, Wagner M, Schüller C, Strauss J. Labuda R, et al. Sci Rep. 2024 Oct 25;14(1):25310. doi: 10.1038/s41598-024-75510-1. Sci Rep. 2024. PMID: 39455635 Free PMC article. - Specific targeting of the KRAS mutational landscape in myeloma as a tool to unveil the elicited antitumor activity.
Sacco A, Federico C, Todoerti K, Ziccheddu B, Palermo V, Giacomini A, Ravelli C, Maccarinelli F, Bianchi G, Belotti A, Ribolla R, Favasuli V, Revenko AS, Macleod AR, Willis B, Cai H, Hauser J, Rooney C, Willis SE, Martin PL, Staniszewska A, Ambrose H, Hanson L, Cattaneo C, Tucci A, Rossi G, Ronca R, Neri A, Mitola S, Bolli N, Presta M, Moschetta M, Ross S, Roccaro AM. Sacco A, et al. Blood. 2021 Nov 4;138(18):1705-1720. doi: 10.1182/blood.2020010572. Blood. 2021. PMID: 34077955 Free PMC article. - Drosophila as a toolkit to tackle cancer and its metabolism.
Jiang H, Kimura T, Hai H, Yamamura R, Sonoshita M. Jiang H, et al. Front Oncol. 2022 Aug 25;12:982751. doi: 10.3389/fonc.2022.982751. eCollection 2022. Front Oncol. 2022. PMID: 36091180 Free PMC article. Review. - LKB1 Inactivation Elicits a Redox Imbalance to Modulate Non-small Cell Lung Cancer Plasticity and Therapeutic Response.
Li F, Han X, Li F, Wang R, Wang H, Gao Y, Wang X, Fang Z, Zhang W, Yao S, Tong X, Wang Y, Feng Y, Sun Y, Li Y, Wong KK, Zhai Q, Chen H, Ji H. Li F, et al. Cancer Cell. 2015 May 11;27(5):698-711. doi: 10.1016/j.ccell.2015.04.001. Epub 2015 Apr 30. Cancer Cell. 2015. PMID: 25936644 Free PMC article. - Targeting autophagy for the treatment of liver diseases.
Ni HM, Williams JA, Yang H, Shi YH, Fan J, Ding WX. Ni HM, et al. Pharmacol Res. 2012 Dec;66(6):463-74. doi: 10.1016/j.phrs.2012.07.003. Epub 2012 Jul 31. Pharmacol Res. 2012. PMID: 22871337 Free PMC article. Review.
References
- Shibutani S, Takeshita M, Grollman AP. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature. 1991;349:431–434. - PubMed
- Wakabayashi N, et al. Keap1-null mutation leads to postnatal lethality due to constitutive Nrf2 activation. Nat Genet. 2003;35:238–245. - PubMed
- Venugopal R, Jaiswal AK. Nrf2 and Nrf1 in association with Jun proteins regulate antioxidant response element-mediated expression and coordinated induction of genes encoding detoxifying enzymes. Oncogene. 1998;17:3145–3156. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA128920/CA/NCI NIH HHS/United States
- U01 CA111294/CA/NCI NIH HHS/United States
- P50 CA062924/CA/NCI NIH HHS/United States
- CA106610/CA/NCI NIH HHS/United States
- CA101973/CA/NCI NIH HHS/United States
- K08 CA106610/CA/NCI NIH HHS/United States
- U01 CA105490/CA/NCI NIH HHS/United States
- CA128920/CA/NCI NIH HHS/United States
- CA111294/CA/NCI NIH HHS/United States
- CA62924/CA/NCI NIH HHS/United States
- F32 CA123887/CA/NCI NIH HHS/United States
- U01 CA084291/CA/NCI NIH HHS/United States
- CA084291/CA/NCI NIH HHS/United States
- R01 CA101973-05/CA/NCI NIH HHS/United States
- CA105490/CA/NCI NIH HHS/United States
- CRUK_/Cancer Research UK/United Kingdom
- R01 CA101973/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous