MicroRNA expression aberration as potential peripheral blood biomarkers for schizophrenia - PubMed (original) (raw)
doi: 10.1371/journal.pone.0021635. Epub 2011 Jun 29.
Sung-Liang Yu, Ming H Hsieh, Chun-Houh Chen, Hsuan-Yu Chen, Chun-Chiang Wen, Yung-Hsiang Huang, Po-Chang Hsiao, Chuhsing Kate Hsiao, Chih-Min Liu, Pan-Chyr Yang, Hai-Gwo Hwu, Wei J Chen
Affiliations
- PMID: 21738743
- PMCID: PMC3126851
- DOI: 10.1371/journal.pone.0021635
MicroRNA expression aberration as potential peripheral blood biomarkers for schizophrenia
Chi-Yu Lai et al. PLoS One. 2011.
Abstract
Since brain tissue is not readily accessible, a new focus in search of biomarkers for schizophrenia is blood-based expression profiling of non-protein coding genes such as microRNAs (miRNAs), which regulate gene expression by inhibiting the translation of messenger RNAs. This study aimed to identify potential miRNA signature for schizophrenia by comparing genome-wide miRNA expression profiles in patients with schizophrenia vs. healthy controls. A genome-wide miRNA expression profiling was performed using a Taqman array of 365 human miRNAs in the mononuclear leukocytes of a learning set of 30 cases and 30 controls. The discriminating performance of potential biomarkers was validated in an independent testing set of 60 cases and 30 controls. The expression levels of the miRNA signature were then evaluated for their correlation with the patients' clinical symptoms, neurocognitive performances, and neurophysiological functions. A seven-miRNA signature (hsa-miR-34a, miR-449a, miR-564, miR-432, miR-548d, miR-572 and miR-652) was derived from a supervised classification with internal cross-validation, with an area under the curve (AUC) of receiver operating characteristics of 93%. The putative signature was then validated in the testing set, with an AUC of 85%. Among these miRNAs, miR-34a was differentially expressed between cases and controls in both the learning (P = 0.005) and the testing set (P = 0.002). These miRNAs were differentially correlated with patients' negative symptoms, neurocognitive performance scores, and event-related potentials. The results indicated that the mononuclear leukocyte-based miRNA profiling is a feasible way to identify biomarkers for schizophrenia, and the seven-miRNA signature warrants further investigation.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
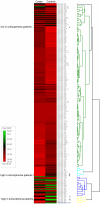
Figure 1. miRNA expression profiling of 221 miRNAs in the learning set of 30 schizophrenia patients and 30 controls.
The heatmaps of individual miRNAs for each group of subjects were presented as the averaged rank-sum of the normalized threshold cycle number, with less numbers indicating higher expression levels. The eight differentially expressed miRNAs were marked by an asterisk.
Figure 2. A comparison of: (A) the TLDA-based miRNA expression levels between the schizophrenia patients (n = 30) and the controls (n = 30) for the seven miRNAs; and (B) quantitative RT-PCR-based miRNA expression levels between the schizophrenia patients (n = 60) and the controls (n = 30) for the same seven miRNAs.
The red, black, and green hues denote relatively high, intermediate, and low expression levels. To emphasize the rank-sum property of the Wilcoxon test, all the expression levels of miRNA were converted to relative expression ranks within each miRNA. Ranks for each miRNA were sorted separately for the control group and disease group before they were placed side-by-side for easier comparison between the two groups and across all putatively informative miRNAs.
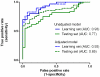
Figure 3. The area under the receiver operative characteristics curve of the seven-miRNA signature in the learning (in blue color) and testing (in green color) sets, respectively, without (in solid line) or with (in dotted line) adjustment for age, gender, education, and tobacco smoking.
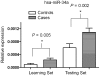
Figure 4. The relations in has-miR-34a miRNA expression levels between two platforms of miRNA quantification, the array-based TLDA vs. individual quantification using quantitative RT-PCR.
The differential expressions in hsa-miR-34a between the cases and controls in both the learning set (P = 0.005) and the testing set (P = 0.002).
Similar articles
- Aberrant expression of microRNAs as biomarker for schizophrenia: from acute state to partial remission, and from peripheral blood to cortical tissue.
Lai CY, Lee SY, Scarr E, Yu YH, Lin YT, Liu CM, Hwang TJ, Hsieh MH, Liu CC, Chien YL, Udawela M, Gibbons AS, Everall IP, Hwu HG, Dean B, Chen WJ. Lai CY, et al. Transl Psychiatry. 2016 Jan 19;6(1):e717. doi: 10.1038/tp.2015.213. Transl Psychiatry. 2016. PMID: 26784971 Free PMC article. - Aberrant microRNA expression in peripheral plasma and mononuclear cells as specific blood-based biomarkers in schizophrenia patients.
Sun XY, Lu J, Zhang L, Song HT, Zhao L, Fan HM, Zhong AF, Niu W, Guo ZM, Dai YH, Chen C, Ding YF, Zhang LY. Sun XY, et al. J Clin Neurosci. 2015 Mar;22(3):570-4. doi: 10.1016/j.jocn.2014.08.018. Epub 2014 Dec 6. J Clin Neurosci. 2015. PMID: 25487174 - Identification of serum microRNAs as diagnostic biomarkers for schizophrenia.
He K, Guo C, Guo M, Tong S, Zhang Q, Sun H, He L, Shi Y. He K, et al. Hereditas. 2019 Jun 27;156:23. doi: 10.1186/s41065-019-0099-3. eCollection 2019. Hereditas. 2019. PMID: 31297041 Free PMC article. - The Role of microRNA in Schizophrenia: A Scoping Review.
Li K, Zhu L, Lv H, Bai Y, Guo C, He K. Li K, et al. Int J Mol Sci. 2024 Jul 12;25(14):7673. doi: 10.3390/ijms25147673. Int J Mol Sci. 2024. PMID: 39062916 Free PMC article. Review. - MiRNAs of peripheral blood as the biomarker of schizophrenia.
He K, Guo C, He L, Shi Y. He K, et al. Hereditas. 2017 Aug 29;155:9. doi: 10.1186/s41065-017-0044-2. eCollection 2018. Hereditas. 2017. PMID: 28860957 Free PMC article. Review.
Cited by
- miRNAs as potential diagnostic biomarkers and pharmacogenomic indicators in psychiatric disorders.
Tsermpini EE, Kalogirou CI, Kyriakopoulos GC, Patrinos GP, Stathopoulos C. Tsermpini EE, et al. Pharmacogenomics J. 2022 Jul;22(4):211-222. doi: 10.1038/s41397-022-00283-7. Epub 2022 Jun 20. Pharmacogenomics J. 2022. PMID: 35725816 Review. - MicroRNAs as biomarkers for CNS disease.
Rao P, Benito E, Fischer A. Rao P, et al. Front Mol Neurosci. 2013 Nov 26;6:39. doi: 10.3389/fnmol.2013.00039. Front Mol Neurosci. 2013. PMID: 24324397 Free PMC article. Review. - Review and Meta-Analyses of TAAR1 Expression in the Immune System and Cancers.
Fleischer LM, Somaiya RD, Miller GM. Fleischer LM, et al. Front Pharmacol. 2018 Jun 26;9:683. doi: 10.3389/fphar.2018.00683. eCollection 2018. Front Pharmacol. 2018. PMID: 29997511 Free PMC article. - microRNAs as novel biomarkers of schizophrenia (Review).
Wang J, Wang Y, Yang J, Huang Y. Wang J, et al. Exp Ther Med. 2014 Dec;8(6):1671-1676. doi: 10.3892/etm.2014.2014. Epub 2014 Oct 9. Exp Ther Med. 2014. PMID: 25371713 Free PMC article. - Increased Expression of Plasma miRNA-320a and let-7b-5p in Heroin-Dependent Patients and Its Clinical Significance.
Liu H, Xu W, Feng J, Ma H, Zhang J, Xie X, Zhuang D, Shen W, Liu H, Zhou W. Liu H, et al. Front Psychiatry. 2021 Jun 29;12:679206. doi: 10.3389/fpsyt.2021.679206. eCollection 2021. Front Psychiatry. 2021. PMID: 34267687 Free PMC article.
References
- Tamminga CA, Holcomb HH. Phenotype of schizophrenia: a review and formulation. Mol Psychiatry. 2005;10:27–39. - PubMed
- van Os J, Kenis G, Rutten BPF. The environment and schizophrenia. Nature. 2010;468:203–212. - PubMed
- Harrison PJ, Weinberger DR. Schizophrenia genes, gene expression, and neuropathology: on the matter of their convergence. Mol Psychiatry. 2005;10:40–68. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical