Distinct roles for classical nuclear import receptors in the growth of multinucleated muscle cells - PubMed (original) (raw)
Distinct roles for classical nuclear import receptors in the growth of multinucleated muscle cells
Monica N Hall et al. Dev Biol. 2011.
Abstract
Proper muscle function is dependent on spatial and temporal control of gene expression in myofibers. Myofibers are multinucleated cells that are formed, repaired and maintained by the process of myogenesis in which progenitor myoblasts proliferate, differentiate and fuse. Gene expression is dependent upon proteins that require facilitated nuclear import, however little is known about the regulation of nucleocytoplasmic transport during the formation of myofibers. We analyzed the role of karyopherin alpha (KPNA), a key classical nuclear import receptor, during myogenesis. We established that five karyopherin alpha paralogs are expressed by primary mouse myoblasts in vitro and that their steady-state levels increase in multinucleated myotubes, suggesting a global increase in demand for classical nuclear import during myogenesis. We used siRNA-mediated knockdown to identify paralog-specific roles for KPNA1 and KPNA2 during myogenesis. KPNA1 knockdown increased myoblast proliferation, whereas KPNA2 knockdown decreased proliferation. In contrast, no proliferation defect was observed with KPNA4 knockdown. Only knockdown of KPNA2 decreased myotube growth. These results identify distinct pathways involved in myoblast proliferation and myotube growth that rely on specific nuclear import receptors suggesting that regulation of classical nuclear import pathways likely plays a critical role in controlling gene expression in skeletal muscle.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures
Fig. 1
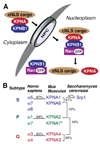
Classical nuclear import. (A) In the cytoplasm, proteins containing a classical nuclear localization sequence (cNLS) are recognized by the classical nuclear import receptor complex consisting of karyopherin alpha (KPNA) and karyopherin beta1 (KPNB1). KPNA recognizes and binds cNLS-containing proteins, while KPNB1 mediates nuclear import of the complex through interactions with the nuclear pore complex (NPC). In the nucleus, KPNB1 is bound by Ran-GTP which induces a conformational change that dissociates the import complex leading to release of the cNLS protein. (B) KPNA paralogs are categorized into three subtypes, S, P and Q. The percent identity between a few subtypes is shown for human, mouse and budding yeast to illustrate the homology between and within subtypes for KPNA paralogs. * The placement of recently discovered mouse KPNA7 into subtype P is tentative (Hu et al., 2010).
Fig. 2
Components of the classical nuclear import system are expressed during myogenesis. (A) Primary mouse myoblasts were differentiated for 0, 24 and 48 h and immunostained for embryonic myosin heavy chain, eMyHC, a marker of differentiation (bar, 50 µm). (B) Total RNA or protein was isolated from primary myoblasts at 0, 24 and 48 h of differentiation. RT-PCR was performed using primers specific for each karyopherin alpha paralog and 18S rRNA as an internal cDNA control. Immunoblotting was performed with antibodies specific for each KPNA paralog or KPNB1 with GAPDH antibody as a loading control. The steady-state levels of karyopherin alpha paralogs increased during myogenesis, whereas KPNB1 levels did not change.
Fig. 3
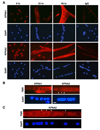
KPNA1 and KPNA2 nuclear steady-state levels differ during myogenesis. (A) Representative fluorescent images of primary myoblasts differentiated for 0, 24 and 48 h and immunostained for either KPNA1 or KPNA2 and counterstained with DAPI (bar, 50 µm). KPNA1 and KPNA2 were detected in all cells with greater nuclear fluorescence observed for KPNA2 at all stages of myogenesis. B) Representative fluorescent images of myotubes at 48 h immunostained for either KPNA1 or KPNA2 (TMR, tetramethylrhodamine) and counterstained with DAPI (bar, 10 µm). KPNA2 appeared to be more highly concentrated in some myonuclei (arrowhead) compared to other myonuclei (arrow), whereas no significant difference in the fluorescence intensity of KPNA1 was observed among nuclei. (C) Confocal z-stacked images of myotubes at 48 h immunostained for KPNA2 (TMR, tetramethylrhodamine) and counterstained with DAPI (bar, 10 µm). Fluorescence was captured in successive confocal slices through DAPI stained nuclei to compare KPNA2 immunostaining among nuclei. Confocal microscopy confirmed that KPNA2 was increased in some myonuclei (arrowhead) compared to other myonuclei (arrow).
Fig. 4
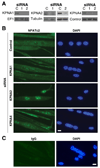
Knockdown of KPNA paralogs alters the steady-state localization of NFATc2. (A) Primary myoblasts were transfected with either control scrambled siRNA or siRNAs directed against KPNA1, KPNA2 or KPNA4. Immunoblotting was performed with antibodies specific for each KPNA to determine the efficiency of knockdown. Antibodies targeting EF1-alpha, alpha-tubulin or a non-specific band (control) obtained during KPNA4 immunoblotting were used as a loading control. (B) Representative fluorescent images of primary myoblasts transfected with different siRNAs. Myoblasts were differentiated for 48 h and immunostained using antibodies targeting (B) NFATc2 or (C) IgG isotype control antibodies and counterstained with DAPI (bar, 25 µm). NFACTc2 was increased in the nuclei of cells treated with KPNA2 and KPNA4 siRNA compared to control with the greatest increase observed with KPNA2 siRNA; no differences were observed with KPNA1 siRNA.
Fig. 5
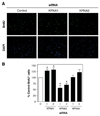
KPNA paralogs have distinct roles in myoblast proliferation. (A) Primary myoblasts were transfected with either control scrambled siRNA or siRNAs (1 or 2) directed against KPNA1, KPNA2 or KPNA4. Cultures were labeled with BrdU and subsequently immunostained. Representative fluorescent images of BrdU immunostaining and nuclear counterstaining with DAPI are shown (bar, 100 µm). (B) Knockdown of KPNA1 significantly increased the percentage of BrdU+ cells while loss of KPNA2 significantly reduced the percentage of BrdU+ cells. No significant difference was observed in cells treated with KPNA4 siRNA. Data are mean ± SEM from three independent experiments, *p<0.05.
Fig. 6
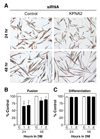
Knockdown of KPNA2 decreases myotube size. (A) Primary myoblasts were transfected with either control scrambled siRNA or one of two siRNAs (1 or 2) against KPNA2. Cells were differentiated for either 24 or 48 h in differentiation media (DM) and immunostained for eMyHC, (bar, 50 µm). (B) The fusion index was significantly decreased with KPNA2 siRNAs at both 24 and 48 h of differentiation. (C) The differentiation index was not altered with KPNA2 siRNA. Data are mean ± SEM from three independent experiments, *p<0.05.
Fig. 7
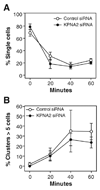
Cell–cell adhesion is not altered following knockdown of KPNA2. (A) A suspension based assay was used to examine cell–cell adhesion over a 60-minute time course. No significant difference in either the percentage of single cells or (B) the percentage of cell clusters with >5 cells was observed with KPNA2 knockdown. Data are mean ± SEM from three independent experiments.
Fig. 8
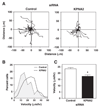
KPNA2 knockdown decreases muscle cell migration. (A) The migration of myocytes transfected with either control scrambled siRNA or siRNA against KPNA2 was analyzed using time-lapse microscopy. Twenty individual cell traces are shown for each siRNA condition. Myocytes treated with KPNA2 siRNA migrated shorter distances than control myocytes. (B) Histogram of the distribution of velocities for control or KPNA2 siRNA treated myocytes. Cells depleted of KPNA2 migrated more slowly than control myocytes. Data are mean ± SEM of 80 cells (20 cells from each of four independent isolates). (C) The mean velocity of migration was reduced in KPNA2 siRNA treated cells compared to control. Data are mean ± SEM of 80 cells (20 cells from each of four independent isolates), *p<0.05.
Similar articles
- Karyopherin Alpha 1 Regulates Satellite Cell Proliferation and Survival by Modulating Nuclear Import.
Choo HJ, Cutler A, Rother F, Bader M, Pavlath GK. Choo HJ, et al. Stem Cells. 2016 Nov;34(11):2784-2797. doi: 10.1002/stem.2467. Epub 2016 Aug 1. Stem Cells. 2016. PMID: 27434733 Free PMC article. - Non-equivalence of nuclear import among nuclei in multinucleated skeletal muscle cells.
Cutler AA, Jackson JB, Corbett AH, Pavlath GK. Cutler AA, et al. J Cell Sci. 2018 Feb 5;131(3):jcs207670. doi: 10.1242/jcs.207670. J Cell Sci. 2018. PMID: 29361530 Free PMC article. - Differential regulation of karyopherin alpha 2 expression by TGF-beta1 and IFN-gamma in normal human epidermal keratinocytes: evident contribution of KPNA2 for nuclear translocation of IRF-1.
Umegaki N, Tamai K, Nakano H, Moritsugu R, Yamazaki T, Hanada K, Katayama I, Kaneda Y. Umegaki N, et al. J Invest Dermatol. 2007 Jun;127(6):1456-64. doi: 10.1038/sj.jid.5700716. Epub 2007 Jan 25. J Invest Dermatol. 2007. PMID: 17255955 - HuR and myogenesis: being in the right place at the right time.
von Roretz C, Beauchamp P, Di Marco S, Gallouzi IE. von Roretz C, et al. Biochim Biophys Acta. 2011 Sep;1813(9):1663-7. doi: 10.1016/j.bbamcr.2011.01.036. Epub 2011 Feb 20. Biochim Biophys Acta. 2011. PMID: 21315776 Review. - Regulation of Skeletal Muscle Myoblast Differentiation and Proliferation by Pannexins.
Langlois S, Cowan KN. Langlois S, et al. Adv Exp Med Biol. 2017;925:57-73. doi: 10.1007/5584_2016_53. Adv Exp Med Biol. 2017. PMID: 27518505 Review.
Cited by
- Cell Surface Proteins for Enrichment and In Vitro Characterization of Human Pluripotent Stem Cell-Derived Myogenic Progenitors.
Tey SR, Mueller M, Reilly M, Switalski C, Robertson S, Sakanaka-Yokoyama M, Suzuki M. Tey SR, et al. Stem Cells Int. 2022 Feb 24;2022:2735414. doi: 10.1155/2022/2735414. eCollection 2022. Stem Cells Int. 2022. PMID: 35251185 Free PMC article. - Karyopherin Alpha 1 Regulates Satellite Cell Proliferation and Survival by Modulating Nuclear Import.
Choo HJ, Cutler A, Rother F, Bader M, Pavlath GK. Choo HJ, et al. Stem Cells. 2016 Nov;34(11):2784-2797. doi: 10.1002/stem.2467. Epub 2016 Aug 1. Stem Cells. 2016. PMID: 27434733 Free PMC article. - Quantitative proteomics reveals regulation of karyopherin subunit alpha-2 (KPNA2) and its potential novel cargo proteins in nonsmall cell lung cancer.
Wang CI, Chien KY, Wang CL, Liu HP, Cheng CC, Chang YS, Yu JS, Yu CJ. Wang CI, et al. Mol Cell Proteomics. 2012 Nov;11(11):1105-22. doi: 10.1074/mcp.M111.016592. Epub 2012 Jul 25. Mol Cell Proteomics. 2012. PMID: 22843992 Free PMC article. - Putting things in place for fertilization: discovering roles for importin proteins in cell fate and spermatogenesis.
Loveland KL, Major AT, Butler R, Young JC, Jans DA, Miyamoto Y. Loveland KL, et al. Asian J Androl. 2015 Jul-Aug;17(4):537-44. doi: 10.4103/1008-682X.154310. Asian J Androl. 2015. PMID: 25994647 Free PMC article. Review. - Importin α: functions as a nuclear transport factor and beyond.
Oka M, Yoneda Y. Oka M, et al. Proc Jpn Acad Ser B Phys Biol Sci. 2018;94(7):259-274. doi: 10.2183/pjab.94.018. Proc Jpn Acad Ser B Phys Biol Sci. 2018. PMID: 30078827 Free PMC article. Review.
References
- Allen DL, Roy RR, Edgerton VR. Myonuclear domains in muscle adaptation and disease. Muscle & Nerve. 1999;22:1350–1360. - PubMed
- Asally M, Yasuda Y, Oka M, Otsuka S, Yoshimura SH, Takeyasu K, Yoneda Y. Nup358, a nucleoporin, functions as a key determinant of the nuclear pore complex structure remodeling during skeletal myogenesis. FEBS Journal. 2011;278:610–621. - PubMed
- Bakkar N, Guttridge DC. NF-kappaB signaling: a tale of two pathways in skeletal myogenesis. Physiological Reviews. 2010;90:495–511. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AR052730/AR/NIAMS NIH HHS/United States
- R01 AR051372-04/AR/NIAMS NIH HHS/United States
- R01 AR051372-05/AR/NIAMS NIH HHS/United States
- R01 AR051372/AR/NIAMS NIH HHS/United States
- R01 AR052730-05/AR/NIAMS NIH HHS/United States
- AR052730/AR/NIAMS NIH HHS/United States
- R01 AR051372-02/AR/NIAMS NIH HHS/United States
- R01 AR052730-04/AR/NIAMS NIH HHS/United States
- AR051372/AR/NIAMS NIH HHS/United States
- R01 AR051372-03/AR/NIAMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Miscellaneous