Structural variants of IFNα preferentially promote antiviral functions - PubMed (original) (raw)
Structural variants of IFNα preferentially promote antiviral functions
Nancy Vázquez et al. Blood. 2011.
Abstract
IFNα, a cytokine with multiple functions in innate and adaptive immunity and a potent inhibitor of HIV, exerts antiviral activity, in part, by enhancing apolipoprotein B mRNA-editing enzyme-catalytic polypeptide-like 3 (APOBEC3) family members. Although IFNα therapy is associated with reduced viral burden, this cytokine also mediates immune dysfunction and toxicities. Through detailed mapping of IFNα receptor binding sites, we generated IFNα hybrids and mutants and determined that structural changes in the C-helix alter the ability of IFN to limit retroviral activity. Selective IFNα constructs differentially block HIV replication and their directional magnitude of inhibition correlates with APOBEC3 levels. Importantly, certain mutants exhibited reduced toxicity as reflected by induced indoleamine 2,3-dioxygenase (IDO), suggesting discreet and shared intracellular signaling pathways. Defining IFN structure and function relative to APOBEC and other antiviral genes may enable design of novel IFN-related molecules preserving beneficial antiviral roles while minimizing negative effects.
Figures
Figure 1
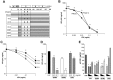
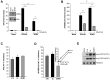
IFNα constructs and antiviral activity. (A) Diagram illustrating construction of IFNα hybrids and mutants. HY1 and HY2 were generated by combining amino acids (aa)1-75 (HY1) or aa1-95 (HY2) from the amino terminus of IFNα21b and carboxy terminus from IFNα2c. Parental IFNα2c tyrosines at aa86 and 90 were present in HY1, while HY2 contains serine and asparagine from IFNα21b. HY4 was generated using HY2 and inserting aa75-81 from IFNα2c retaining serine (aa86) and asparagine (aa90) from IFNα21b. SDM1 differs from HY4 only at aa86 containing a tyrosine. SDM2 derived from HY4 has serine and tyrosine at position 86 and 90, respectively. CM3 differs from SDM1 only at position 86 (S86K). (B) Macrophages (4 × 105) were exposed to HIV for 2 hours, washed and treated once with the indicated concentrations of IFNα (1, 0.1, 0.01, 0.001 ng/mL) parental molecules (*P < .003) or (C) IFN hybrids (HY1, HY2, and HY4 at 0.1-0.1ng/mL *P < .05). Viral replication on day 10 after infection as determined by p24 viral Ag (n = 3). (D) Macrophages (4 × 105) were exposed to HIV for 2 hours, washed, and treated once with SDM1, SDM2, and CM3 (0.01-0.1ng/mL) or media. Ten-day supernatants were tested for p24 by ELISA. Representative donor in duplicates (n = 3). Mutant IFNα molecules compared with cultures infected with HIV in the absence of IFN 2-tailed t test, **P < .001 or *P < .01. SDM1 and SDM2 at 0.1 or 0.01ng/mL compared with CM3 at 0.1 (P < .001) or 0.01 ng/mL (P < .05). (E) Activated (anti-CD3/CD28) T lymphocytes were exposed to HIV IIIB and then incubated in the presence or absence of parental IFN or mutants. (1.0-0.01 ng/mL). *P < .01 HIV alone vs IFN constructs. **P < .05 HIV alone vs CM3. IFNα21b, SDM1, and SDM2 (at 0.1 and 0.01ng/mL) were more effective in controlling HIV replication compared with CM3, P < .02. Data represent mean + SEM.
Figure 2
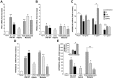
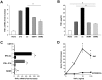
Antiviral gene induction by parental IFNα, hybrid, and mutants. After treatment of macrophages (6 × 106) with IFNα21b, IFNα2c, their hybrids or mutants (0.1 ng/mL) for 4 hours, total mRNA was isolated and examined for (A) PKR and (B) TRIM22 gene transcription using real-time PCR. Representative donor; n = 3. For PKR 2-tailed t test for IFNα-treated compared with control cells (None), **P < .01 (SDM1, SDM2, IFNα21b, IFNα2c) and *P < .05 (CM3, HY4). For TRIM22, *P < .05 for IFN constructs compared with untreated macrophages. Induction of PKR and TRIM22 by SDM1, SDM2, or parental IFN is not significantly different from that of CM3. (C) APOBEC3G gene expression in macrophages incubated with IFN constructs at 0.1-10 ng/mL. *P < .03 untreated vs IFN-treated constructs, *P < .03 parental IFN vs SDM1, **P < .05 IFNα21b vs SDM1. (D) APOBEC3G gene expression correlates with antiretroviral activity. Macrophages (6 × 106) were treated in parallel with IFNα mutants, hybrid (HY4), or parental IFNα (1 ng/mL) for 4 hours and mRNA analyzed for APOBEC3G by RT-PCR. IFNα21b, IFNα2c, SDM1, and SDM2 significantly enhance APOBEC3G gene expression. **P < .001. For HY4 and CM3, *P < .01. APOBEC3G induction by SDM1, SDM2, IFNα2c, and IFNα21b is statistically higher than CM3, *P < .01. Representative donor, n = 3. (E) Macrophages were examined for APOBEC3A transcription after IFNα treatment (1 ng/mL). IFNα parent, hybrid, and mutants induced APOBEC3A (*P < .01), with maximal levels induced by mutant SDM1, *P < .01. Parental IFN vs SDM1, P < .05 and SDM1 vs CM3, **P = .01. Inset: APOBEC3A gene expression in monocytes exposed to IFN constructs. Untreated compared with IFN-treated constructs, *P < .05. Representative donor in duplicate n = 3. Data represent mean + SEM.
Figure 3
IFNα constructs 3-D models. IFN homology models for parental IFN (IFNα2c and IFNα21b), hybrids, and mutants were generated as described in “Homology modeling.” The ribbon diagram renderings of IFN homology models are colored by secondary structure (pink = α helix, blue = 310 helix, cyan = turn, white = coil). Mutations in the C-loop of the molecule in amino acid positions 86 and 90 are shown in red and green, respectively. Overall the structures are very similar with the mean root mean square deviation (RMSD) between the C-α carbons being 2.5 Å (median RMSD = 2.4 Å). Most of the structural differences between the models occur in loop regions, specifically positions 21-31, 45-51, and 100-110. Docking of IFN-binding ectodomain of the homology model of IFNα2c to IFNAR2 is shown (top left panel). Yellow color on IFNα2c model illustrates IFNAR2 interacting region (top right panel). Coloring on secondary structure was performed using Visual Molecular Dynamics (VMD) program.
Figure 4
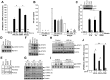
IFNα and mutant signaling. (A) Macrophages were pretreated or not with anti-IFNAR (CD118) for 20 minutes and treated with parental IFNα or SDM1 for 4 hours. Levels of APOBEC3A for SDM1 compared with parental IFNs *P < .01. APOBEC3A transcription by RT-PCR shows that blockade of IFNAR2 reduces APOBEC3A gene expression, **P < .01. (B) Macrophages were exposed to HIV 2 hours, washed and incubated with CD118 antibody or isotype control for 20 minutes. IFNα21b, IFNα2c SDM1 were then added once at 1 ng/mL and supernatants collected at day 13 after infection and tested for p24 (*P < .01, HIV alone vs IFN-treated). (C) Macrophages were treated with 1 ng/mL of IFNα21b, IFNα2c, or SDM1 in the presence or absence of excess soluble IFNAR2. *P < .05 SDM1 vs 21B and 2C. Inset: Whole cell lysates from macrophages treated with IFNα2c, SDM1 and SDM2 were analyzed for P-JAK1 by Western blot. Representative assay n = 2. (D) Whole cell protein lysates collected after 15-minute exposure to IFNα (0.1 ng/mL) were examined for STAT activation. Levels of STAT1 phosphorylation were analyzed by Western blot using an anti-pSTAT1 antibody. Membranes were then probed with STAT1 antibody to confirm equivalent protein loading. Representative data, n = 4. (E) STAT2 activation was detected 15 minutes after IFN (1 ng/mL) by immunoblotting using pSTAT2 and STAT2 antibodies or β-actin (n = 3). (F) Enhanced phosphorylation of STAT3, STAT5, and STAT6 after treatment with indicated IFNα constructs (1 ng/mL) for 20 minutes (n = 3). (G) Whole cell lysates from macrophages exposed to medium alone, IFNα2c, SDM1, and SDM2 for the indicated time points were analyzed by Western blot for pSTAT1 (Y), pSTAT3 (Y), pSTAT3 (S), pSTAT6 (Y) and tubulin. (H) Macrophages were treated with JAK inhibitor I for 1 hour before addition of IFNα and cell lysates analyzed for pSTAT1 or (I) pSTAT3 by Western blot. Interference with IFNAR-associated JAK suppresses STAT1 and STAT3 phosphorylation when compared with macrophages not treated with inhibitor. Representative data, n = 3. (J) Total RNA analyzed by RT-PCR for APOBEC3A transcription in macrophages pretreated with JAK inhibitor 1 hour prior to 4-hour IFNα. Inhibition of JAK almost completely abrogates APOBEC3A transcription. Representative data, n = 3, *P < .003. Levels of APOBEC3A for SDM1 compared with parental IFNs, **P < .01. Data represent mean + SEM.
Figure 5
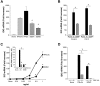
Comparison of IFN constructs for induction of IDO. (A) Total mRNA was isolated from macrophages (6 × 106) treated with IFNα21b, IFNα2c, SDM1 or SDM2. SDM1 (S86- > Y) and SDM2 (N90- > Y) induced reduced IDO gene expression in comparison to parental IFN molecules at 1 ng/mL (*P < .02). Representative data, n = 3. (B) Total mRNA analyses of macrophages incubated in the presence or absence of Polymyxin B and exposed to IFNα2c or SDM1 as indicated for 4 hours (*P < .05). (C) Real-time PCR of monocytes treated with indicated concentrations of IFNα2c, SDM1 for 4 hours confirms limited IDO transcription for mutant IFN, *P < .01. Inset: CD118 antibody blocks parental IFN and SDM1 IDO gene induction. Representative data, n = 3, *P < .01 IFNα2c vs SDM1. *P < .01 IFN alone compared with anti-CD118/IFN-treated. (D) Real-time PCR analyses comparing levels of transcriptional activity for IDO in macrophages cultures pretreated with JAK inhibitor (100nM) 60 minutes before 4-hour treatment with 1 ng/mL of IFNα2c or SDM1. *P < .01 for levels of IDO for SDM1 vs IFNα2c and IFN constructs versus inhibitor/IFN-treated. Data represent mean + SEM.
Figure 6
Regulation of IFNα-induced APOBEC3 and IDO by NFκB and calmodulin/CaMK. (A) Whole cell protein lysates collected after treatment with parental IFN or SDM1 (1 ng/mL, 20 minutes) were examined for IκBα phosphorylation by Western blot using an anti–P-IκBα antibody (inset). Transcription of IDO mRNA in macrophages that were pretreated with Bay 11-7082 (10μM) for 1 hour followed by 4 hours of parental or SDM1 mutant is diminished by 82% and 96%, respectively *P < .05 for IDO levels 2C compared with SDM1 and IFN treated compared with inhibitor/IFN treated. Representative data, n = 3. (B) Transcription of APOBEC3A in cells that were pretreated with Bay 11-7082 inhibitor for 1 hour followed by IFNα2c or SDM1 (4 hours), *P < .05 for control compared with IFN-treated; IFNα2c vs SDM1; and IFN constructs treated versus inhibitor/IFN-treated. Representative data, n = 3. (C, D) Macrophage cultures were pretreated for 1 hour with the calmodulin inhibitor W7 (40μM) or (D inset) CaMKII inhibitor KN93 (10-40μM) before 4-hour treatment with parental IFNs and SDM1 mutant. Total mRNA was examined for APOBEC3A and IDO gene transcription, in C P = not significant; in D *P < .05. (E) Whole cell lysates were analyzed for pSTAT1(Y701)(S727) and tubulin (30 minutes). Representative data, n = 3.
Figure 7
Differential TNFα induction after exposure to IFN constructs. (A) Macrophages were examined for TNFα gene expression (*P < .003 SDM1 and SDM2 vs IFNα2c; P < .02 SDM1 and SDM2 vs IFNα21b (4 hours) by RT-PCR. P < .002 untreated vs IFN constructs treated macrophages. (B) Supernatant TNFα protein (4 hours) was determined by ELISA (*P < .001 IFN constructs compared with untreated and SDM1 or SDM2 vs parental IFNα2c; mutants vs IFNα21b P < .02). Representative data n = 3. (C) Total mRNA was isolated from macrophages that were incubated with anti-CD118 before treatment with parental IFNs or SDM1 mutant for 4 hours. TNFα gene transcription was examined by RT-PCR. (*P < .01; ns, not significant; P < .01 for parental IFN vs SDM1). (D) Macrophages were treated with rhTNFα (1-100 ng/mL) for 4 hours and total mRNA examined for IDO or APOBEC3A gene expression by RT-PCR. Data represent mean + SEM * P < .05, untreated compared with TNFα-treated.
Similar articles
- RIG-I activation inhibits HIV replication in macrophages.
Wang Y, Wang X, Li J, Zhou Y, Ho W. Wang Y, et al. J Leukoc Biol. 2013 Aug;94(2):337-41. doi: 10.1189/jlb.0313158. Epub 2013 Jun 6. J Leukoc Biol. 2013. PMID: 23744645 Free PMC article. - IFN-α induces APOBEC3G, F, and A in immature dendritic cells and limits HIV-1 spread to CD4+ T cells.
Mohanram V, Sköld AE, Bächle SM, Pathak SK, Spetz AL. Mohanram V, et al. J Immunol. 2013 Apr 1;190(7):3346-53. doi: 10.4049/jimmunol.1201184. Epub 2013 Feb 20. J Immunol. 2013. PMID: 23427247 - Interferon-alpha mediates restriction of human immunodeficiency virus type-1 replication in primary human macrophages at an early stage of replication.
Cheney KM, McKnight Á. Cheney KM, et al. PLoS One. 2010 Oct 20;5(10):e13521. doi: 10.1371/journal.pone.0013521. PLoS One. 2010. PMID: 20975956 Free PMC article. - Antiviral roles of APOBEC proteins against HIV-1 and suppression by Vif.
Romani B, Engelbrecht S, Glashoff RH. Romani B, et al. Arch Virol. 2009;154(10):1579-88. doi: 10.1007/s00705-009-0481-y. Epub 2009 Aug 12. Arch Virol. 2009. PMID: 19669862 Review. - APOBEC deaminases as cellular antiviral factors: a novel natural host defense mechanism.
Franca R, Spadari S, Maga G. Franca R, et al. Med Sci Monit. 2006 May;12(5):RA92-8. Med Sci Monit. 2006. PMID: 16641889 Review.
Cited by
- Impact of APOBEC3s on the occurrence, development and prognosis of esophageal squamous cell carcinoma.
Yang F, Xiao H, Dai X, Xu M, Li M, Bai J, Dai N. Yang F, et al. Future Oncol. 2025 Jan;21(1):117-125. doi: 10.1080/14796694.2024.2442300. Epub 2024 Dec 30. Future Oncol. 2025. PMID: 39840662 Free PMC article. Review. - Clustered and genome-wide transient mutagenesis in human cancers: Hypermutation without permanent mutators or loss of fitness.
Roberts SA, Gordenin DA. Roberts SA, et al. Bioessays. 2014 Apr;36(4):382-393. doi: 10.1002/bies.201300140. Epub 2014 Feb 26. Bioessays. 2014. PMID: 24615916 Free PMC article. - IFN-α subtypes: distinct biological activities in anti-viral therapy.
Gibbert K, Schlaak JF, Yang D, Dittmer U. Gibbert K, et al. Br J Pharmacol. 2013 Mar;168(5):1048-58. doi: 10.1111/bph.12010. Br J Pharmacol. 2013. PMID: 23072338 Free PMC article. Review. - Interferon Alpha Subtype-Specific Suppression of HIV-1 Infection In Vivo.
Lavender KJ, Gibbert K, Peterson KE, Van Dis E, Francois S, Woods T, Messer RJ, Gawanbacht A, Müller JA, Münch J, Phillips K, Race B, Harper MS, Guo K, Lee EJ, Trilling M, Hengel H, Piehler J, Verheyen J, Wilson CC, Santiago ML, Hasenkrug KJ, Dittmer U. Lavender KJ, et al. J Virol. 2016 Jun 10;90(13):6001-6013. doi: 10.1128/JVI.00451-16. Print 2016 Jul 1. J Virol. 2016. PMID: 27099312 Free PMC article. - Gene therapy with plasmids encoding IFN-β or IFN-α14 confers long-term resistance to HIV-1 in humanized mice.
Abraham S, Choi JG, Ortega NM, Zhang J, Shankar P, Swamy NM. Abraham S, et al. Oncotarget. 2016 Nov 29;7(48):78412-78420. doi: 10.18632/oncotarget.12512. Oncotarget. 2016. PMID: 27729616 Free PMC article.
References
- Richman DD, Margolis DM, Delaney M, Greene WC, Hazuda D, Pomerantz RJ. The challenge of finding a cure for HIV infection. Science. 2009;323(5919):1304–1307. - PubMed
- Orenstein JM, Fox C, Wahl SM. Macrophages as a source of HIV during opportunistic infections. Science. 1997;276(5320):1857–1861. - PubMed
- Wahl SM, Greenwell-Wild T, Peng G, Ma G, Orenstein JM, Vazquez N. Viral and host cofactors facilitate HIV-1 replication in macrophages. J Leukoc Biol. 2003;74(5):726–735. - PubMed
- Brass AL, Dykxhoorn DM, Benita Y, et al. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319(5865):921–926. - PubMed
- Bishop KN, Holmes RK, Sheehy AM, Malim MH. APOBEC-mediated editing of viral RNA. Science. 2004;305(5684):645. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials