Crystal structure of menin reveals binding site for mixed lineage leukemia (MLL) protein - PubMed (original) (raw)
Crystal structure of menin reveals binding site for mixed lineage leukemia (MLL) protein
Marcelo J Murai et al. J Biol Chem. 2011.
Abstract
Menin is a tumor suppressor protein that is encoded by the MEN1 (multiple endocrine neoplasia 1) gene and controls cell growth in endocrine tissues. Importantly, menin also serves as a critical oncogenic cofactor of MLL (mixed lineage leukemia) fusion proteins in acute leukemias. Direct association of menin with MLL fusion proteins is required for MLL fusion protein-mediated leukemogenesis in vivo, and this interaction has been validated as a new potential therapeutic target for development of novel anti-leukemia agents. Here, we report the first crystal structure of menin homolog from Nematostella vectensis. Due to a very high sequence similarity, the Nematostella menin is a close homolog of human menin, and these two proteins likely have very similar structures. Menin is predominantly an α-helical protein with the protein core comprising three tetratricopeptide motifs that are flanked by two α-helical bundles and covered by a β-sheet motif. A very interesting feature of menin structure is the presence of a large central cavity that is highly conserved between Nematostella and human menin. By employing site-directed mutagenesis, we have demonstrated that this cavity constitutes the binding site for MLL. Our data provide a structural basis for understanding the role of menin as a tumor suppressor protein and as an oncogenic co-factor of MLL fusion proteins. It also provides essential structural information for development of inhibitors targeting the menin-MLL interaction as a novel therapeutic strategy in MLL-related leukemias.
Figures
FIGURE 1.
Crystal structure of the menin homolog from N. vectensis. A, sequence alignment of Nematostella (Nema) and human (Men1) menins. Color coding highlights identical (blue) and conserved (yellow) residues. Positions of secondary structure elements derived from the crystal structure are shown and colored following the schemes in B and C. The regions deleted in Nematostella menin for crystallization are labeled del1 and del2; the three long loops are labeled L1, L2, and L3. Residues lining putative MLL binding site are in red boxes. B, the overall fold of menin is composed of three TRP motifs (cyan), which are flanked by N-terminal (purple) and C-terminal (blue) helical bundles and covered by a three-stranded antiparallel β-sheet (green). C, the structure of Nematostella menin rotated by 90°. Helices are shown as cylinders, and the TRP motifs are numbered. D, the central cavity in Nematostella menin. The residues lining the cavity are shown in surface representation (pale green). E, representation of conserved residues based on multiple sequence alignment of 21 menin homologs prepared using the ConSurf server (37).
FIGURE 2.
Homology model of human menin. A, a model of full-length human menin shown in surface representation. Loops are labeled L1, L2, and L3; NLS1 is shown in magenta, NLS2 is shown in red, and the central cavity is shown in green. B, electrostatic potential calculated for the homology model of human menin.
FIGURE 3.
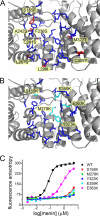
The central cavity in menin constitutes the MLL MBM1 binding site. A, side chains forming the central cavity in human menin are shown in blue. Residues that differ between the human and Nematostella homologs are labeled and shown in red. B, residues mutated in human menin to disrupt MLL binding are labeled and shown in cyan. C, fluorescence polarization binding assay demonstrating decrease of MLL MBM1 binding affinity to menin mutants. Kd value for wild type menin is 77 ± 38 n
m
and 812 ± 305 n
m
for E363K mutant. Four remaining menin mutants bind MBM1 very weakly with Kd > 10 μ
m
.
FIGURE 4.
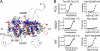
Characterization of MEN1 missense mutations. A, distribution of MEN1 missense mutations mapped onto the homology model of human menin. Buried and solvent exposed residues mutated in MEN1 are shown in blue and red, respectively. B, comparison of thermodynamic stability and MLL binding affinity for P12L, A242V, and wild type human menin.
Similar articles
- Structural insights into inhibition of the bivalent menin-MLL interaction by small molecules in leukemia.
Shi A, Murai MJ, He S, Lund G, Hartley T, Purohit T, Reddy G, Chruszcz M, Grembecka J, Cierpicki T. Shi A, et al. Blood. 2012 Nov 29;120(23):4461-9. doi: 10.1182/blood-2012-05-429274. Epub 2012 Aug 30. Blood. 2012. PMID: 22936661 Free PMC article. - The menin tumor suppressor protein is an essential oncogenic cofactor for MLL-associated leukemogenesis.
Yokoyama A, Somervaille TC, Smith KS, Rozenblatt-Rosen O, Meyerson M, Cleary ML. Yokoyama A, et al. Cell. 2005 Oct 21;123(2):207-18. doi: 10.1016/j.cell.2005.09.025. Cell. 2005. PMID: 16239140 - Molecular basis of the mixed lineage leukemia-menin interaction: implications for targeting mixed lineage leukemias.
Grembecka J, Belcher AM, Hartley T, Cierpicki T. Grembecka J, et al. J Biol Chem. 2010 Dec 24;285(52):40690-8. doi: 10.1074/jbc.M110.172783. Epub 2010 Oct 20. J Biol Chem. 2010. PMID: 20961854 Free PMC article. - Recent Progress of Small Molecule Menin-MLL Interaction Inhibitors as Therapeutic Agents for Acute Leukemia.
Lei H, Zhang SQ, Fan S, Bai HR, Zhao HY, Mao S, Xin M. Lei H, et al. J Med Chem. 2021 Nov 11;64(21):15519-15533. doi: 10.1021/acs.jmedchem.1c00872. Epub 2021 Nov 2. J Med Chem. 2021. PMID: 34726905 Review. - Challenges and opportunities in targeting the menin-MLL interaction.
Cierpicki T, Grembecka J. Cierpicki T, et al. Future Med Chem. 2014 Mar;6(4):447-62. doi: 10.4155/fmc.13.214. Future Med Chem. 2014. PMID: 24635524 Free PMC article. Review.
Cited by
- PepSite: prediction of peptide-binding sites from protein surfaces.
Trabuco LG, Lise S, Petsalaki E, Russell RB. Trabuco LG, et al. Nucleic Acids Res. 2012 Jul;40(Web Server issue):W423-7. doi: 10.1093/nar/gks398. Epub 2012 May 16. Nucleic Acids Res. 2012. PMID: 22600738 Free PMC article. - KAT8 Regulates Androgen Signaling in Prostate Cancer Cells.
Kim JY, Yu J, Abdulkadir SA, Chakravarti D. Kim JY, et al. Mol Endocrinol. 2016 Aug;30(8):925-36. doi: 10.1210/me.2016-1024. Epub 2016 Jun 7. Mol Endocrinol. 2016. PMID: 27268279 Free PMC article. - Detection of disordered regions in globular proteins using ¹³C-detected NMR.
Gray FL, Murai MJ, Grembecka J, Cierpicki T. Gray FL, et al. Protein Sci. 2012 Dec;21(12):1954-60. doi: 10.1002/pro.2174. Protein Sci. 2012. PMID: 23047544 Free PMC article. - Structural insights into inhibition of the bivalent menin-MLL interaction by small molecules in leukemia.
Shi A, Murai MJ, He S, Lund G, Hartley T, Purohit T, Reddy G, Chruszcz M, Grembecka J, Cierpicki T. Shi A, et al. Blood. 2012 Nov 29;120(23):4461-9. doi: 10.1182/blood-2012-05-429274. Epub 2012 Aug 30. Blood. 2012. PMID: 22936661 Free PMC article. - Menin and Menin-Associated Proteins Coregulate Cancer Energy Metabolism.
Chou CW, Tan X, Hung CN, Lieberman B, Chen M, Kusi M, Mitsuya K, Lin CL, Morita M, Liu Z, Chen CL, Huang TH. Chou CW, et al. Cancers (Basel). 2020 Sep 22;12(9):2715. doi: 10.3390/cancers12092715. Cancers (Basel). 2020. PMID: 32971831 Free PMC article.
References
- Chandrasekharappa S. C., Guru S. C., Manickam P., Olufemi S. E., Collins F. S., Emmert-Buck M. R., Debelenko L. V., Zhuang Z., Lubensky I. A., Liotta L. A., Crabtree J. S., Wang Y., Roe B. A., Weisemann J., Boguski M. S., Agarwal S. K., Kester M. B., Kim Y. S., Heppner C., Dong Q., Spiegel A. M., Burns A. L., Marx S. J. (1997) Science 276, 404–407 - PubMed
- Marx S. J. (2005) Nat. Rev. Cancer 5, 367–375 - PubMed
- Agarwal S. K., Guru S. C., Heppner C., Erdos M. R., Collins R. M., Park S. Y., Saggar S., Chandrasekharappa S. C., Collins F. S., Spiegel A. M., Marx S. J., Burns A. L. (1999) Cell 96, 143–152 - PubMed
- Heppner C., Bilimoria K. Y., Agarwal S. K., Kester M., Whitty L. J., Guru S. C., Chandrasekharappa S. C., Collins F. S., Spiegel A. M., Marx S. J., Burns A. L. (2001) Oncogene 20, 4917–4925 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources