A RING E3-substrate complex poised for ubiquitin-like protein transfer: structural insights into cullin-RING ligases - PubMed (original) (raw)
A RING E3-substrate complex poised for ubiquitin-like protein transfer: structural insights into cullin-RING ligases
Matthew F Calabrese et al. Nat Struct Mol Biol. 2011.
Abstract
How RING E3 ligases mediate E2-to-substrate ubiquitin-like protein (UBL) transfer remains unknown. Here we address how the RING E3 RBX1 positions NEDD8's E2 (UBC12) and substrate (CUL1). We find that existing structures are incompatible with CUL1 NEDD8ylation and report a new conformation of RBX1 that places UBC12 adjacent to CUL1. We propose RING domain rotation as a general mechanism for UBL transfer for the largest family of E3s.
Figures
Figure 1
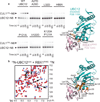
UBC12–RBX1 interactions. (a) WT and mutant UBC12-mediated CUL1 NEDD8ylation in vitro. Mutated residues are orange on UBC12 (otherwise cyan)–RBX1 (pink) model. (b) 2D HSQC [15N-1H] spectra of 15N-UBC12core in the absence (red) and presence (blue) of equimolar RBX1RING. A subset of shifted resonances are labeled in the spectra. Residues with chemical shift differences greater than one standard deviation above the mean are labeled and colored orange on UBC12 (otherwise cyan)–RBX1 (pink) model.
Figure 2
Models from prior structures reveal an E2-to-substrate gap. (a) Structural model of CUL1CTD–RBX1–UBC12core (colored light green, pink, cyan respectively) complex based on previous CUL1–RBX1 and RING–E2 structures–,, (Supplementary Fig. 1). CUL1 NEDD8 modification site and UBC12 catalytic cysteine are indicated by blue and yellow spheres respectively. (b) In vitro CUL1 NEDD8ylation assays using disulfide-linked (S~S) and unlinked (SH SH) split’n’coexpress CUL1–RBX1 (ref. 7). Pairs A–C refer to three distinct combinations of engineered cysteines (Supplementary Fig. 2).
Figure 3
Structure of CUL1CTD–RBX1 in new conformation. (a) One copy of CUL1CTD (dark green) –RBX1 (purple) from the asymmetric unit. (b) Same as (a), but rotated ~70° in y and ~20° in x. (c) Structural overlay with previous CUL1CTD (light green) oriented similarly to (a),. (d) Structural overlay with previous RBX1 (pink) in same orientation as (b). (e) Structural model of CUL1CTD–RBX1–UBC12core (light green, purple, cyan respectively) complex based on the new RBX1 conformation. (f) Bis-Maleimidoethane (BMOE)-crosslinking between engineered cysteine mutants of Ubc12 and CUL1CTD. Residues mutated to cysteine are numbered on the model (left, view rotated by ~70° and ~40° about the×and z-axes relative to panel e) and products of crosslinking reactions are shown for all possible Ubc12+CUL1–RBX1 combinations (right). Note residue numbering for yeast Ubc12. For reference, Ubc12 catalytic cysteine and CUL1 NEDD8ylation site are shown as sticks. (g) In vitro CUL1 NEDD8ylation for UBC12 mutants at the predicted interface. Mutated residues are shown on the model (left).
Comment in
- Conformational flexibility and rotation of the RING domain in activation of cullin-RING ligases.
Rahighi S, Dikic I. Rahighi S, et al. Nat Struct Mol Biol. 2011 Aug 3;18(8):863-5. doi: 10.1038/nsmb.2117. Nat Struct Mol Biol. 2011. PMID: 21811311 No abstract available.
Similar articles
- Structure of a RING E3 trapped in action reveals ligation mechanism for the ubiquitin-like protein NEDD8.
Scott DC, Sviderskiy VO, Monda JK, Lydeard JR, Cho SE, Harper JW, Schulman BA. Scott DC, et al. Cell. 2014 Jun 19;157(7):1671-84. doi: 10.1016/j.cell.2014.04.037. Cell. 2014. PMID: 24949976 Free PMC article. - A dual E3 mechanism for Rub1 ligation to Cdc53.
Scott DC, Monda JK, Grace CR, Duda DM, Kriwacki RW, Kurz T, Schulman BA. Scott DC, et al. Mol Cell. 2010 Sep 10;39(5):784-96. doi: 10.1016/j.molcel.2010.08.030. Mol Cell. 2010. PMID: 20832729 Free PMC article. - Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation.
Duda DM, Borg LA, Scott DC, Hunt HW, Hammel M, Schulman BA. Duda DM, et al. Cell. 2008 Sep 19;134(6):995-1006. doi: 10.1016/j.cell.2008.07.022. Cell. 2008. PMID: 18805092 Free PMC article. - NEDD8 and ubiquitin ligation by cullin-RING E3 ligases.
Baek K, Scott DC, Schulman BA. Baek K, et al. Curr Opin Struct Biol. 2021 Apr;67:101-109. doi: 10.1016/j.sbi.2020.10.007. Epub 2020 Nov 5. Curr Opin Struct Biol. 2021. PMID: 33160249 Free PMC article. Review. - Genetically engineered mouse models for functional studies of SKP1-CUL1-F-box-protein (SCF) E3 ubiquitin ligases.
Zhou W, Wei W, Sun Y. Zhou W, et al. Cell Res. 2013 May;23(5):599-619. doi: 10.1038/cr.2013.44. Epub 2013 Mar 26. Cell Res. 2013. PMID: 23528706 Free PMC article. Review.
Cited by
- Building and remodelling Cullin-RING E3 ubiquitin ligases.
Lydeard JR, Schulman BA, Harper JW. Lydeard JR, et al. EMBO Rep. 2013 Dec;14(12):1050-61. doi: 10.1038/embor.2013.173. Epub 2013 Nov 15. EMBO Rep. 2013. PMID: 24232186 Free PMC article. Review. - DCNL1 functions as a substrate sensor and activator of cullin 2-RING ligase.
Heir P, Sufan RI, Greer SN, Poon BP, Lee JE, Ohh M. Heir P, et al. Mol Cell Biol. 2013 Apr;33(8):1621-31. doi: 10.1128/MCB.01342-12. Epub 2013 Feb 11. Mol Cell Biol. 2013. PMID: 23401859 Free PMC article. - Structural insights into the catalysis and regulation of E3 ubiquitin ligases.
Buetow L, Huang DT. Buetow L, et al. Nat Rev Mol Cell Biol. 2016 Oct;17(10):626-42. doi: 10.1038/nrm.2016.91. Epub 2016 Aug 3. Nat Rev Mol Cell Biol. 2016. PMID: 27485899 Free PMC article. Review. - Proteasome lid bridges mitochondrial stress with Cdc53/Cullin1 NEDDylation status.
Bramasole L, Sinha A, Gurevich S, Radzinski M, Klein Y, Panat N, Gefen E, Rinaldi T, Jimenez-Morales D, Johnson J, Krogan NJ, Reis N, Reichmann D, Glickman MH, Pick E. Bramasole L, et al. Redox Biol. 2019 Jan;20:533-543. doi: 10.1016/j.redox.2018.11.010. Epub 2018 Nov 17. Redox Biol. 2019. PMID: 30508698 Free PMC article. - Twists and turns in ubiquitin-like protein conjugation cascades.
Schulman BA. Schulman BA. Protein Sci. 2011 Dec;20(12):1941-54. doi: 10.1002/pro.750. Epub 2011 Nov 9. Protein Sci. 2011. PMID: 22012881 Free PMC article. Review.
References
- Deshaies RJ, Joazeiro CA. Annu Rev Biochem. 2009;78:399–434. - PubMed
- Zheng N, Wang P, Jeffrey PD, Pavletich NP. Cell. 2000;102:533–539. - PubMed
- Mace PD, et al. J Biol Chem. 2008;283:31633–31640. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA082491-10/CA/NCI NIH HHS/United States
- R01 GM069530-10/GM/NIGMS NIH HHS/United States
- P41 RR015301/RR/NCRR NIH HHS/United States
- P30 CA021765-33S1/CA/NCI NIH HHS/United States
- R01 GM069530-09/GM/NIGMS NIH HHS/United States
- RR-15301/RR/NCRR NIH HHS/United States
- R01CA082491/CA/NCI NIH HHS/United States
- R01 CA082491-08/CA/NCI NIH HHS/United States
- P30 CA021765/CA/NCI NIH HHS/United States
- P30 CA021765-31/CA/NCI NIH HHS/United States
- R01 CA082491/CA/NCI NIH HHS/United States
- HHMI_/Howard Hughes Medical Institute/United States
- P30 CA021765-32/CA/NCI NIH HHS/United States
- R01 GM069530/GM/NIGMS NIH HHS/United States
- R01 GM069530-08/GM/NIGMS NIH HHS/United States
- R01GM069530/GM/NIGMS NIH HHS/United States
- R01 CA082491-09/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous