Embryonic lethality in mice lacking the nuclear factor of activated T cells 5 protein due to impaired cardiac development and function - PubMed (original) (raw)
Embryonic lethality in mice lacking the nuclear factor of activated T cells 5 protein due to impaired cardiac development and function
Man Chi Mak et al. PLoS One. 2011.
Abstract
Nuclear factor of activated T cells 5 protein (NFAT5) is thought to be important for cellular adaptation to osmotic stress by regulating the transcription of genes responsible for the synthesis or transport of organic osmolytes. It is also thought to play a role in immune function, myogenesis and cancer invasion. To better understand the function of NFAT5, we developed NFAT5 gene knockout mice. Homozygous NFAT5 null (NFAT5(-/-)) mouse embryos failed to develop normally and died after 14.5 days of embryonic development (E14.5). The embryos showed peripheral edema, and abnormal heart development as indicated by thinner ventricular wall and reduced cell density at the compact and trabecular areas of myocardium. This is associated with reduced level of proliferating cell nuclear antigen and increased caspase-3 in these tissues. Cardiomyocytes from E14.5 NFAT5(-/-) embryos showed a significant reduction of beating rate and abnormal Ca(2+) signaling profile as a consequence of reduced sarco(endo)plasmic reticulum Ca(2+)-ATPase (SERCA) and ryanodine receptor (RyR) expressions. Expression of NFAT5 target genes, such as HSP 70 and SMIT were reduced in NFAT5(-/-) cardiomyocytes. Our findings demonstrated an essential role of NFAT5 in cardiac development and Ca(2+) signaling. Cardiac failure is most likely responsible for the peripheral edema and death of NFAT5(-/-) embryos at E14.5 days.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
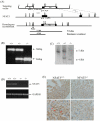
Figure 1. Targeting vector to generate NFAT5 null mutant mice.
(A) The genomic structure of mouse NFAT5 gene. Exons are represented in black boxes and numbered with 1–16. Introns are represented by lines. Targeting construct for NFAT5 knockout which contain the thymidine kinase gene (tk) and the neomycin resistance gene (Neo). The targeting vector was linearized with Not1 restriction endonuclease. Exon 5 and 6 are shown in boxes and the restriction map of the endogenous NFAT5 and the mutant allele after homologous recombination are shown. The primers (ptkR, pKB2.5T7R, p3.pA and p3RA) used for PCR screening are shown as arrows. For Southern hybridization screening, 600-bp Spe1-BamH1 fragment was used as external probe, the expected sizes of EcoRV fragments from wild type allele and homologous recombinant were 7.5 kb and 4.5 kb respectively. E: EcoRV, B: BamH1, K:Kpn1, N:Not1, S:Spe1 (B) Deletion of NFAT5 gene was verified by genotyping using PCR and (C) Southern blot analyses. (D) RT-PCR analysis of NFAT5 RNA in hearts from E14.5 wild-type and NFAT5−/− embryos. (E) Representative photomicrography showing expression of NFAT5 in ventricular compact zone in E14.5 NFAT5+/+ embryonic hearts (red arrows), n = 5.
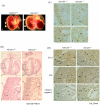
Figure 2. Peripheral edema in embryos at E14.5d.
(A) Edema is apparent in the E14.5 NFAT5−/− embryos (b) compared with a wild-type littermate (a). The arrow indicate the outer skin layer. (B) Histological comparison of NFAT5+/+ (a and c) and NFAT5−/− (b and d) embryonic heart at 14.5 days. (C) Representative photomicrograph showing proliferating cell nuclear antigen (PCNA)-positive cells (arrows) in compact zone and trabecular region in E14.5 NFAT5+/+ and NFAT5−/− hearts. (D) Representative photomicrography showing the (a and b) Bcl-2, (c and d) Bax and (e and f) cleaved caspase-3 staining (arrows) on E14.4 NFAT5+/+and NFAT5−/− hearts. Com, compact layer; E, endocardial cushion; Endo, endocardium; Ep, epicardium; LV, left ventricle; Tr, trabecular. n = 5 Scale bar = 50 µm.
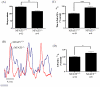
Figure 3. Beating rate of cardiomyocytes from E14.5 embryos.
(A) The beating rate is presented in total counts of beating in cardiomyocytes per minute. Data presented as mean ± S.E.M. **P<0.01, one-way ANOVA. (B) Ca2+ records from a rested (not paced) NFAT5+/+ and NFAT5−/− cardiomyocytes. Effect of NFAT5 on amplitude of [Ca2+]i transient (C), time to peak (D) and time to 50% decay (t50) in single ventricular cardiomyocytes. Data presented as mean ± S.E.M. ≥13 cardiomyocytes were measured from each individual animal. NFAT5+/+, n = 6; NFAT5−/−, n = 5 *P<0.05; ***P<0.0001, student t-test.
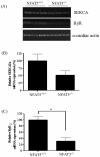
Figure 4. Effect of NFAT5 on the mRNA expression of RyR and SERCA in embryonic cardiomyocytes.
(A) Representative semi-quantitative RT-PCR showing the relative mRNA abundance of SERCA and RyR in NFAT5−/− cardiomyocytes compared to wild-type control. (B) Histogram showing the relative quantification of SERCA of semi-quantitative RT-PCR. (C) Histogram showing the relative quantification of RyR of semi-quantitative RT-PCR. Data presented as mean ± S.E.M. **P<0.05, student t-test, n = 3.
Similar articles
- T-tubule disruption promotes calcium alternans in failing ventricular myocytes: mechanistic insights from computational modeling.
Nivala M, Song Z, Weiss JN, Qu Z. Nivala M, et al. J Mol Cell Cardiol. 2015 Feb;79:32-41. doi: 10.1016/j.yjmcc.2014.10.018. Epub 2014 Nov 6. J Mol Cell Cardiol. 2015. PMID: 25450613 Free PMC article. - Loss of AKAP150 promotes pathological remodelling and heart failure propensity by disrupting calcium cycling and contractile reserve.
Li L, Li J, Drum BM, Chen Y, Yin H, Guo X, Luckey SW, Gilbert ML, McKnight GS, Scott JD, Santana LF, Liu Q. Li L, et al. Cardiovasc Res. 2017 Feb;113(2):147-159. doi: 10.1093/cvr/cvw221. Epub 2016 Nov 17. Cardiovasc Res. 2017. PMID: 27856611 Free PMC article. - Further study on the role of HSP70 on Ca2+ homeostasis in rat ventricular myocytes subjected to simulated ischemia.
Liu J, Kam KW, Borchert GH, Kravtsov GM, Ballard HJ, Wong TM. Liu J, et al. Am J Physiol Cell Physiol. 2006 Feb;290(2):C583-91. doi: 10.1152/ajpcell.00145.2005. Epub 2005 Oct 5. Am J Physiol Cell Physiol. 2006. PMID: 16207797 - Regulation of sarco(endo)plasmic reticulum Ca2+-ATPase and calsequestrin gene expression in the heart.
Zarain-Herzberg A, Estrada-Avilés R, Fragoso-Medina J. Zarain-Herzberg A, et al. Can J Physiol Pharmacol. 2012 Aug;90(8):1017-28. doi: 10.1139/y2012-057. Epub 2012 Jul 11. Can J Physiol Pharmacol. 2012. PMID: 22784385 Review. - Altered sarco(endo)plasmic reticulum calcium adenosine triphosphatase 2a content: Targets for heart failure therapy.
Liu G, Li SQ, Hu PP, Tong XY. Liu G, et al. Diab Vasc Dis Res. 2018 Jul;15(4):322-335. doi: 10.1177/1479164118774313. Epub 2018 May 15. Diab Vasc Dis Res. 2018. PMID: 29762054 Review.
Cited by
- Not enough by half: NFAT5 haploinsufficiency in two patients with Epstein-Barr virus susceptibility.
Lopez-Rivera DO, Castano-Jaramillo LM, Yamazaki-Nakashimada MA, Ramirez Uribe RMN, Corcuera Delgado CT, Ignorosa-Arellano KR, Medina-Torres EA, Berrón Ruiz L, Espinosa-Padilla SE, Scheffler-Mendoza SC, López-Velázquez G, Cruz-Munoz ME, Lugo Reyes SO. Lopez-Rivera DO, et al. Front Immunol. 2022 Sep 27;13:959733. doi: 10.3389/fimmu.2022.959733. eCollection 2022. Front Immunol. 2022. PMID: 36238298 Free PMC article. Review. - A Novel Single-Nucleotide Polymorphism in W NT4 Promoter Affects Its Transcription and Response to FSH in Chicken Follicles.
Zhong C, Wang Y, Liu C, Jiang Y, Kang L. Zhong C, et al. Genes (Basel). 2022 Oct 1;13(10):1774. doi: 10.3390/genes13101774. Genes (Basel). 2022. PMID: 36292659 Free PMC article. - Tonicity-independent regulation of the osmosensitive transcription factor TonEBP (NFAT5).
Halterman JA, Kwon HM, Wamhoff BR. Halterman JA, et al. Am J Physiol Cell Physiol. 2012 Jan 1;302(1):C1-8. doi: 10.1152/ajpcell.00327.2011. Epub 2011 Oct 12. Am J Physiol Cell Physiol. 2012. PMID: 21998140 Free PMC article. Review. - Potential Role of Gene Regulator NFAT5 in the Pathogenesis of Diabetes Mellitus.
Cen L, Xing F, Xu L, Cao Y. Cen L, et al. J Diabetes Res. 2020 Sep 15;2020:6927429. doi: 10.1155/2020/6927429. eCollection 2020. J Diabetes Res. 2020. PMID: 33015193 Free PMC article. Review. - Isoform-Selective NFAT Inhibitor: Potential Usefulness and Development.
Kitamura N, Kaminuma O. Kitamura N, et al. Int J Mol Sci. 2021 Mar 8;22(5):2725. doi: 10.3390/ijms22052725. Int J Mol Sci. 2021. PMID: 33800389 Free PMC article. Review.
References
- Rushlow C, Warrior R. The rel family of proteins. Bioessays. 1992;14:89–95. - PubMed
- Rao A, Luo C, Hogan PG. Transcription factors of the NFAT family: regulation and function. Annu Rev Immunol. 1997;15:707–747. - PubMed
- Dahl SC, Handler JS, Kwon HM. Hypertonicity-induced phosphorylation and nuclear localization of the transcription factor TonEBP. Am J Physiol Cell Physiol. 2001;280:C248–253. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous