Enzymatic basis of ribosomal peptide prenylation in cyanobacteria - PubMed (original) (raw)
. 2011 Aug 31;133(34):13698-705.
doi: 10.1021/ja205458h. Epub 2011 Aug 4.
Affiliations
- PMID: 21766822
- PMCID: PMC3170831
- DOI: 10.1021/ja205458h
Enzymatic basis of ribosomal peptide prenylation in cyanobacteria
John A McIntosh et al. J Am Chem Soc. 2011.
Abstract
The enzymatic basis of ribosomal peptide natural product prenylation has not been reported. Here, we characterize a prenyltransferase, LynF, from the TruF enzyme family. LynF is the first characterized representative of the TruF protein family, which is responsible for both reverse- and forward-O-prenylation of tyrosine, serine, and threonine in cyclic peptides known as cyanobactins. We show that LynF reverse O-prenylates tyrosine in macrocyclic peptides. Based upon these results, we propose that the TruF family prenylates mature cyclic peptides, from which the leader sequence and other enzyme recognition elements have been excised. This differs from the common model of ribosomal peptide biosynthesis, in which a leader sequence is required to direct post-translational modifications. In addition, we find that reverse O-prenylated tyrosine derivatives undergo a facile Claisen rearrangement at 'physiological' temperature in aqueous buffers, leading to forward C-prenylated products. Although the Claisen rearrangement route to natural products has been chemically anticipated for at least 40 years, it has not been demonstrated as a route to prenylated natural products. Here, we show that the Claisen rearrangement drives phenolic C-prenylation in at least one case, suggesting that this route should be reconsidered as a mechanism for the biosynthesis of prenylated phenolic compounds.
Figures
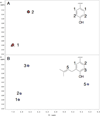
Figure 1. Prenylation in natural products
(A) Representative cyanobactin peptide natural products, showing groups derived from DMAPP in blue. (B) Two possible enzymatic mechanisms of phenol _ortho_-C-prenylation. First, DMAPP is dephosphorylated to yield a cation that can react either at oxygen (pathway I) or at carbon (pathyway II). In principle, a Claisen rearrangement from pathway I could then yield the C-prenylated product. Only pathway II has been previously linked to enzymatic modification.
Figure 2. Defining the biosynthetic route to prenylated cyanobactins
Proposed biosynthetic scheme for lyn pathway showing modification of precursor peptide by heterocyclization, proteolysis, macrocyclization and prenylation. The first steps in this route are supported by previous enzymological studies, but the timing of prenylation was not known. Enzyme recognition elements are highlighted. Analogues (3–6) were used to assess prenylation of each possible biosynthetic intermediate and are shown in analogous colors beneath each proposed biosynthetic intermediate. Pro was substituted as an approximate isostere for thiazole in later analogues. No reaction was observed with any intermediate except the final cyclic peptide, showing that prenylation occurs at a late step, after all enzyme recognition elements have been excised.
Figure 3. LynF is _ortho_-C-prenylating
The aromatic / olefin region of the HSQC NMR spectra are shown for boc-tyrosine (14) (A) and its purified enzymatic product with LynF (B), clearly indicating a single, forward C-prenylation event. Similar spectra were also observed for reactions containing 6. NMR and MS characterization of compounds is presented in Supporting Figures S1, S3, and S5.
Figure 4. LynF catalyzes reverse O-prenylation of tyrosine
The aromatic / olefin region of 1H NMR spectra are shown (A) for boc-tyrosine (14) (B) the HPLC-purified intermediate LynF product (31), and (C) the final reaction product (30). These spectra clearly indicate that the first product of the LynF reaction is reverse-O-prenyl tyrosine, which subsequently rearranges to give the C-prenylated product.
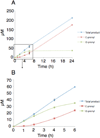
Figure 5. Time course of boc-L-Tyr (14) reaction followed by HPLC
The reaction was performed in quadruplicate, with variation indicated by error bars (A), (B). Initial product of the reaction is almost exclusively O-prenylated as shown at 1 h, 2 h, and 4 h time points (B). After 4 h, the level of O-prenylated intermediate reaches steady-state and its levels are constant through 24 h, accompanied by steady increase in the concentration of C-prenylated final product 29. This allowed kinetic constants for the Claisen rearrangement to be directly determined, since at steady state the concentration of the O-prenyl intermediate can be assumed to be a constant.
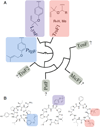
Figure 6. Phylogenetic analysis of the TruF / LynF family
(A) TruF-group proteins cluster according to whether natural products are prenylated (top) or non-prenylated (bottom). Chemical products are shown for each TruF-like protein, where they exist. No TruF-like sequence relatives can be identified outside of cyanobactin gene clusters using either BLAST searching or sequence alignments with other PT family proteins, indicating that this is a novel group of PTs. (B) Actual product structures of prenylagaramide (left) and trunkamide (right) pathways; predicted enzymatic product of lyn pathway (middle) prior to Claisen rearrangement.
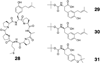
Scheme 1. NMR characterized products
Scheme 2. Claisen rearrangement pathway
Similar articles
- Pericyclic prenylation: peptide modification through a Claisen rearrangement.
Majmudar JD, Gibbs RA. Majmudar JD, et al. Chembiochem. 2011 Dec 16;12(18):2723-6. doi: 10.1002/cbic.201100612. Epub 2011 Nov 24. Chembiochem. 2011. PMID: 22114066 - Genome-Mining-Based Discovery of the Cyclic Peptide Tolypamide and TolF, a Ser/Thr Forward O-Prenyltransferase.
Purushothaman M, Sarkar S, Morita M, Gugger M, Schmidt EW, Morinaka BI. Purushothaman M, et al. Angew Chem Int Ed Engl. 2021 Apr 6;60(15):8460-8465. doi: 10.1002/anie.202015975. Epub 2021 Mar 5. Angew Chem Int Ed Engl. 2021. PMID: 33586286 Free PMC article. - Aromatic Claisen Rearrangements of O-prenylated tyrosine and model prenyl aryl ethers: Computational study of the role of water on acceleration of Claisen rearrangements.
Osuna S, Kim S, Bollot G, Houk KN. Osuna S, et al. European J Org Chem. 2013 May 1;2013(14):10.1002/ejoc.201201738. doi: 10.1002/ejoc.201201738. European J Org Chem. 2013. PMID: 24376368 Free PMC article. - Cyanobactins-ribosomal cyclic peptides produced by cyanobacteria.
Sivonen K, Leikoski N, Fewer DP, Jokela J. Sivonen K, et al. Appl Microbiol Biotechnol. 2010 May;86(5):1213-25. doi: 10.1007/s00253-010-2482-x. Epub 2010 Feb 27. Appl Microbiol Biotechnol. 2010. PMID: 20195859 Free PMC article. Review. - Catalysts for the Enzymatic Lipidation of Peptides.
Zheng Y, Cong Y, Schmidt EW, Nair SK. Zheng Y, et al. Acc Chem Res. 2022 May 3;55(9):1313-1323. doi: 10.1021/acs.accounts.2c00108. Epub 2022 Apr 20. Acc Chem Res. 2022. PMID: 35442036 Free PMC article. Review.
Cited by
- Genome Mining for New Enzyme Chemistry.
Nguyen DT, Mitchell DA, van der Donk WA. Nguyen DT, et al. ACS Catal. 2024 Mar 12;14(7):4536-4553. doi: 10.1021/acscatal.3c06322. eCollection 2024 Apr 5. ACS Catal. 2024. PMID: 38601780 Free PMC article. Review. - Structure Prediction and Genome Mining-Aided Discovery of the Bacterial C-Terminal Tryptophan Prenyltransferase PalQ.
Miyata A, Ito S, Fujinami D. Miyata A, et al. Adv Sci (Weinh). 2024 Feb;11(6):e2307372. doi: 10.1002/advs.202307372. Epub 2023 Dec 7. Adv Sci (Weinh). 2024. PMID: 38059776 Free PMC article. - Engineering lanthipeptides by introducing a large variety of RiPP modifications to obtain new-to-nature bioactive peptides.
Fu Y, Xu Y, Ruijne F, Kuipers OP. Fu Y, et al. FEMS Microbiol Rev. 2023 May 19;47(3):fuad017. doi: 10.1093/femsre/fuad017. FEMS Microbiol Rev. 2023. PMID: 37096385 Free PMC article. Review. - Chemoenzymatic Late-Stage Modifications Enable Downstream Click-Mediated Fluorescent Tagging of Peptides.
Colombano A, Dalponte L, Dall'Angelo S, Clemente C, Idress M, Ghazal A, Houssen WE. Colombano A, et al. Angew Chem Int Ed Engl. 2023 Apr 11;62(16):e202215979. doi: 10.1002/anie.202215979. Epub 2023 Mar 10. Angew Chem Int Ed Engl. 2023. PMID: 36815722 Free PMC article. - Emulating nonribosomal peptides with ribosomal biosynthetic strategies.
Mordhorst S, Ruijne F, Vagstad AL, Kuipers OP, Piel J. Mordhorst S, et al. RSC Chem Biol. 2022 Dec 6;4(1):7-36. doi: 10.1039/d2cb00169a. eCollection 2023 Jan 4. RSC Chem Biol. 2022. PMID: 36685251 Free PMC article. Review.
References
- Brandt W, Bräuer L, Günnewich N, Kufka J, Rausch F, Schulze D, Schulze E, Weber R, Zakharova S, Wessjohann L. Phytochemistry. 2009;70:1758–1775. - PubMed
- Nguyen UT, Goody RS, Alexandrov K. ChemBioChem. 2010;11:1194–1201. - PubMed
- Gibbs RA. Nat. Chem. Biol. 2005;1:7–8. - PubMed
- Casey PJ, Seabra MC. J. Biol. Chem. 1996;271:5289–5292. - PubMed
- Sacchettini JC, Poulter CD. Science. 1997;277:1788–1789. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- GM071425/GM/NIGMS NIH HHS/United States
- R01 GM071425-05/GM/NIGMS NIH HHS/United States
- R01 GM071425/GM/NIGMS NIH HHS/United States
- R01 GM071425-01A1/GM/NIGMS NIH HHS/United States
- R01 GM071425-04/GM/NIGMS NIH HHS/United States
- R01 GM071425-03/GM/NIGMS NIH HHS/United States
- R01 GM071425-02/GM/NIGMS NIH HHS/United States
- R01 GM071425-04S1/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources