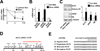Flow-Dependent Regulation of Kruppel-Like Factor 2 Is Mediated by MicroRNA-92a - PubMed (original) (raw)
. 2011 Aug 2;124(5):633-41.
doi: 10.1161/CIRCULATIONAHA.110.005108. Epub 2011 Jul 18.
Han Xiao, Andrés Laguna-Fernandez, Guadalupe Villarreal Jr, Kuei-Chun Wang, Greg G Geary, Yuzhi Zhang, Wei-Chi Wang, Hsien-Da Huang, Jing Zhou, Yi-Shuan Li, Shu Chien, Guillermo Garcia-Cardena, John Y-J Shyy
Affiliations
- PMID: 21768538
- PMCID: PMC3511909
- DOI: 10.1161/CIRCULATIONAHA.110.005108
Flow-Dependent Regulation of Kruppel-Like Factor 2 Is Mediated by MicroRNA-92a
Wei Wu et al. Circulation. 2011.
Abstract
Background: Upregulated by atheroprotective flow, the transcription factor Krüppel-like factor 2 (KLF2) is crucial for maintaining endothelial function. MicroRNAs (miRNAs) are noncoding small RNAs that regulate gene expression at the posttranscriptional level. We examined the role of miRNAs, particularly miR-92a, in the atheroprotective flow-regulated KLF2.
Methods and results: Dicer knockdown increased the level of KLF2 mRNA in human umbilical vein endothelial cells, suggesting that KLF2 is regulated by miRNA. In silico analysis predicted that miR-92a could bind to the 3' untranslated region of KLF2 mRNA. Overexpression of miR-92a decreased the expression of KLF2 and the KLF2-regulated endothelial nitric oxide synthase and thrombomodulin at mRNA and protein levels. A complementary finding is that miR-92a inhibitor increased the mRNA and protein expression of KLF2, endothelial nitric oxide synthase, and thrombomodulin. Subsequent studies revealed that atheroprotective laminar flow downregulated the level of miR-92a precursor to induce KLF2, and the level of this flow-induced KLF2 was reduced by miR-92a precursor. Furthermore, miR-92a level was lower in human umbilical vein endothelial cells exposed to the atheroprotective pulsatile shear flow than under atheroprone oscillatory shear flow. Anti-Ago1/2 immunoprecipitation coupled with real-time polymerase chain reaction revealed that pulsatile shear flow decreased the functional targeting of miR-92a precursor/KLF2 mRNA in human umbilical vein endothelial cells. Consistent with these findings, mouse carotid arteries receiving miR-92a precursor exhibited impaired vasodilatory response to flow.
Conclusions: Atheroprotective flow patterns decrease the level of miR-92a, which in turn increases KLF2 expression to maintain endothelial homeostasis.
Figures
Figure 1. miRNAs are involved in the regulation of KLF2 mRNA
(A) HUVECs were subjected to laminar flow (12 dyn/cm2) for 6 hr. After the addition of DRB (2 µg/ml), then the cells were continuously exposed to laminar flow or static conditions for additional 2 and 4 hr. The levels of KLF2 and GAPDH mRNAs were measured by qRT-PCR, and the KLF2/GAPDH mRNA ratio is plotted as a percentage of that in the untreated static cells. (B,C) HUVECs were transfected with 20 nM Dicer siRNA or control RNA for 48 hr. qRT-PCR (B) and Western blot analysis (C) were performed to detect the mRNA levels of KLF2 and eNOS and protein levels of KLF2, eNOS, and TM, respectively. Histone H1 served as the internal control of nuclear extracts. The bar graphs are mean±SD from 3 independent experiments. * p<0.05 between Dicer siRNA and control RNA, analyzed by ANOVA followed by Dunnett’s test (A) or Student t test (B,C). (D) The miRNA binding sites in the KLF2 3’UTR are predicted by bioinformatics algorithms. (E) The seed region of miR-92a and its target sequences at the KLF2 3’UTR of several mammalian species: Homo sapiens, Pan troglodytes, Mus Musculus, and Rattus Norvegicus
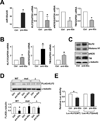
Figure 2. miR-92a targets KLF2 mRNA and decreases KLF2 translation
HUVECs were transfected with 20 nM miR-92a precursor (pre-92a) or control RNA. At 48 hr, the levels of miR-92a relative to U6 RNA and KLF2, eNOS, and TM mRNA as ratios to GAPDH were assessed by qRT-PCR. In (B) and (C), HUVECs were transfected with 20 nM miR-92a inhibitor (anti-92a) or control RNA. The mRNA levels of KLF2, eNOS, and TM as ratios to GAPDH were assessed by qRT-PCR. T h e protein levels of KLF2, Histone H1 and eNOS were determined by Western blotting. (D) HEK293 cells were transfected with the wild-type FLAGKLF2 (WT), FLAG-KLF2 (mut) (miR-92a binding site mutation), or Flag-KLF2 (Δ) (miR-92a 24 binding site deletion) together with 20 nM pre-92a or control RNA for 48 hr. The cells were then lysed, and the level of exogenously expressed FLAG-KLF2 fusion proteins was detected by Western blot analysis with anti-FLAG. Shown in the bottom panel is the densitometry analysis of the protein amount normalized to that in the control RNA-transfected cells. (E) HEK293 cells were transfected with the wild-type Luc-KLF2-3’UTR(WT) or Luc-KLF2-3’UTR(mut) together with 20 nM pre-92a or control RNA and CMV-β-gal. The luciferase activity was normalized to that of β-gal. The data represent mean±SD from 3 independent experiments. * p<0.05 between cells transfected with pre-92a and control RNA.
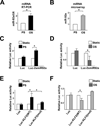
Figure 3. Shear stress-induction of KLF2 is mediated through miR-92a
(A) HUVECs were exposed to laminar flow for 4, 8 or 16 hr. qRT-PCR was performed to detect the level of miR-92a, which was normalized to that of U6 RNA. * p<0.05 compared with static controls (time 0), analyzed by one-way ANOVA followed by Dunnett’s test. (B–F) HUVECs were transfected with 20 nM control RNA or pre-92a for 48 hr and then exposed to laminar flow for 8 hr. (B) KLF2 mRNA level was detected by qRT-PCR and (C) protein level was assessed by Western blot analysis. (D,E) eNOS and TM mRNA levels were detected by qRT-PCR and (F) protein level was assessed by Western blot analysis, and the results of statistical analyses are shown in the right. The data represent mean±SD from 3 independent experiments. * p<0.05 between the indicated groups, analyzed by two-way ANOVA followed by the Bonferroni posthoc test.
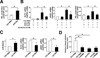
Figure 4. PS down-regulates, but OS up-regulates, miR-92a expression in ECs
(A) HUVECs were exposed to PS (12±4 dyn/cm2) or OS (0±4 dyn/cm2) for 8 hr. qRT-PCR was performed to detect the level of miR-92a, which was normalized to that of U6 RNA. (B) The expression of miR-92a in ECs exposed to PS or OS flow assessed by miRNA microarray. (C,D) BAECs were transfected with Luc-2×miR92 reporter or control plasmid for 24 hr and then exposed to PS or OS flow for 12 hr. The luciferase activity was measured and normalized to β-gal activity. (E, F) BAECs were transfected with Luc-KLF2(WT) or Luc-KLF2(mut) for 24 hr and then exposed to PS or OS flow for 12 hr. The luciferase activity was measured and normalized to β-gal activity. The data represent mean±SD from 3 independent experiments. * p<0.05 between the 2 groups being compared by Student t test (A,B) or two-way ANOVA (C–F) followed by the Bonferroni posthoc test.
Figure 5. miRISC regulates miR-92a
HUVECs were exposed to PS (A,C) or OS (B,D) for 8 hr. The Ago1- or Ago2-associated miRNAs and mRNAs were enriched by IP with the use of anti-Ago1 (A,B) or anti-Ago2 (C,D). mRNA levels of miR-92a and KLF2 were detected by qRT-PCR and normalized to those of Ago1 or Ago2 protein. The data represent mean±SD from 3 independent experiments. * p<0.05 for PS or OS vs. static control, as analyzed by Student t test.
Figure 6. miR-92a regulates endothelial function in vitro and ex vivo
(A) HUVECs were transfected with control RNA or anti-92a. After 48 hr, the NO bioavailability was detected by fluorometric assay and expressed as nmol/106 cells. In (B), HUVECs were transfected with pre-92a and infected with Ad-KLF2-3’UTR or Ad-null for 48 hr. The level of KLF2 and eNOS mRNA was assessed by qRT-PCR and NO bioavailability was measured. (C) pre-92a or control RNA was administered to the carotid arteries of C57BL6 mice by pluronic gel F-127. Five days later, the arteries were isolated. The expression levels of miR-92a, KLF2 and eNOS in the isolated vessels (n=6) were assessed by qRT-PCR. miR-92a level was normalized to that of U6 RNA, whereas KLF2 and eNOS levels were normalized to that of GAPDH. The data represent mean±SD. (D) The flow-induced vasodilation ex vivo was measured by use of the SoftEdge Acquisiton Subsystem in the presence or absence of L-NAME (1 µM). The dilation ability is defined as the percentage of the diameter change of the flow-induced dilation compared to the diameter change of the PE-induced constriction. The bars represent mean±SEM. p<0.05 between the 2 groups being compared by Student t test (A,C) or two-way ANOVA followed by the Bonferroni posthoc test (B,D).
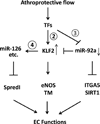
Figure 7. Shear stress regulation of KLF2
The diagram shows the regulatory circuitry of the responses of transcription factors and miRNAs to atheroprotective shear flow. The circled numerals refer to the Table numbers. Shear stress with a forward direction regulates the expressions of KLF2 and miR-92a through several TFs (in Supplemental Tables 2 and 3, respectively). Serving as a transcription factor, KLF2 transactivates the expression of downstream genes such as eNOS and TM. In addition, KLF2 may bind to the promoter region of some miRNAs, including miR-126, to upregulate their transcription directly (Supplemental Table 4). In turn, the network of KLF2 and miRNAs regulates the expression of factors that control anti-inflammatory, anti-thrombotic, anti-proliferative, anti-angiogenic, anti-oxidant, and anti-fibrotic effects to maintain EC functions. The methods for computational analysis are described in the supplements.
Comment in
- Crippling of Krüppel (-like factor 2) by bad flow portends a miRky day for endothelial function.
Irani K. Irani K. Circulation. 2011 Aug 2;124(5):541-3. doi: 10.1161/CIRCULATIONAHA.111.043299. Circulation. 2011. PMID: 21810672 No abstract available.
Similar articles
- Site-specific microRNA-92a regulation of Kruppel-like factors 4 and 2 in atherosusceptible endothelium.
Fang Y, Davies PF. Fang Y, et al. Arterioscler Thromb Vasc Biol. 2012 Apr;32(4):979-87. doi: 10.1161/ATVBAHA.111.244053. Epub 2012 Jan 19. Arterioscler Thromb Vasc Biol. 2012. PMID: 22267480 Free PMC article. - Shear stress sustains atheroprotective endothelial KLF2 expression more potently than statins through mRNA stabilization.
van Thienen JV, Fledderus JO, Dekker RJ, Rohlena J, van Ijzendoorn GA, Kootstra NA, Pannekoek H, Horrevoets AJ. van Thienen JV, et al. Cardiovasc Res. 2006 Nov 1;72(2):231-40. doi: 10.1016/j.cardiores.2006.07.008. Epub 2006 Jul 12. Cardiovasc Res. 2006. PMID: 16945356 - Inhibition of microRNA-92a prevents endothelial dysfunction and atherosclerosis in mice.
Loyer X, Potteaux S, Vion AC, Guérin CL, Boulkroun S, Rautou PE, Ramkhelawon B, Esposito B, Dalloz M, Paul JL, Julia P, Maccario J, Boulanger CM, Mallat Z, Tedgui A. Loyer X, et al. Circ Res. 2014 Jan 31;114(3):434-43. doi: 10.1161/CIRCRESAHA.114.302213. Epub 2013 Nov 19. Circ Res. 2014. PMID: 24255059 - Mechanosensitive microRNAs-role in endothelial responses to shear stress and redox state.
Marin T, Gongol B, Chen Z, Woo B, Subramaniam S, Chien S, Shyy JY. Marin T, et al. Free Radic Biol Med. 2013 Sep;64:61-8. doi: 10.1016/j.freeradbiomed.2013.05.034. Epub 2013 May 30. Free Radic Biol Med. 2013. PMID: 23727269 Free PMC article. Review. - Atheroprotective mechanisms of shear stress-regulated microRNAs.
Boon RA, Hergenreider E, Dimmeler S. Boon RA, et al. Thromb Haemost. 2012 Oct;108(4):616-20. doi: 10.1160/TH12-07-0491. Epub 2012 Sep 5. Thromb Haemost. 2012. PMID: 22955103 Review.
Cited by
- MicroRNA therapeutics for cardiovascular disease: opportunities and obstacles.
van Rooij E, Olson EN. van Rooij E, et al. Nat Rev Drug Discov. 2012 Nov;11(11):860-72. doi: 10.1038/nrd3864. Epub 2012 Oct 19. Nat Rev Drug Discov. 2012. PMID: 23080337 Free PMC article. Review. - Site-specific microRNA-92a regulation of Kruppel-like factors 4 and 2 in atherosusceptible endothelium.
Fang Y, Davies PF. Fang Y, et al. Arterioscler Thromb Vasc Biol. 2012 Apr;32(4):979-87. doi: 10.1161/ATVBAHA.111.244053. Epub 2012 Jan 19. Arterioscler Thromb Vasc Biol. 2012. PMID: 22267480 Free PMC article. - Thrombomodulin and the vascular endothelium: insights into functional, regulatory, and therapeutic aspects.
Martin FA, Murphy RP, Cummins PM. Martin FA, et al. Am J Physiol Heart Circ Physiol. 2013 Jun 15;304(12):H1585-97. doi: 10.1152/ajpheart.00096.2013. Epub 2013 Apr 19. Am J Physiol Heart Circ Physiol. 2013. PMID: 23604713 Free PMC article. Review. - Regulation of nuclear transcription by mitochondrial RNA in endothelial cells.
Sriram K, Qi Z, Yuan D, Malhi NK, Liu X, Calandrelli R, Luo Y, Tapia A, Jin S, Shi J, Salas M, Dang R, Armstrong B, Priceman SJ, Wang PH, Liao J, Natarajan R, Zhong S, Bouman Chen Z. Sriram K, et al. Elife. 2024 Jan 22;13:e86204. doi: 10.7554/eLife.86204. Elife. 2024. PMID: 38251974 Free PMC article. - Fluid Shear Stress Regulates the Landscape of microRNAs in Endothelial Cell-Derived Small Extracellular Vesicles and Modulates the Function of Endothelial Cells.
Chung J, Kim KH, Yu N, An SH, Lee S, Kwon K. Chung J, et al. Int J Mol Sci. 2022 Jan 24;23(3):1314. doi: 10.3390/ijms23031314. Int J Mol Sci. 2022. PMID: 35163238 Free PMC article.
References
- Caro CG, Fitz-Gerald JM, Schroter RC. Atheroma and arterial wall shear. Observation, correlation and proposal of a shear dependent mass transfer mechanism for atherogenesis. Proc R Soc Lond B Biol Sci. 1971;177:109–159. - PubMed
- Zarins CK, Giddens DP, Bharadvaj BK, Sottiurai VS, Mabon RF, Glagov S. Carotid bifurcation atherosclerosis. Quantitative correlation of plaque localization with flow velocity profiles and wall shear stress. Circ Res. 1983;53:502–514. - PubMed
- Ku DN, Giddens DP, Zarins CK, Glagov S. Pulsatile flow and atherosclerosis in the human carotid bifurcation. Positive correlation between plaque location and low oscillating shear stress. Arteriosclerosis. 1985;5:293–302. - PubMed
- Asakura T, Karino T. Flow patterns and spatial distribution of atherosclerotic lesions in human coronary arteries. Circ Res. 1990;66:1045–1066. - PubMed
- Atkins GB, Jain MK. Role of Kruppel-like transcription factors in endothelial biology. Circ Res. 2007;100:1686–1695. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 HL089940-03/HL/NHLBI NIH HHS/United States
- R01 HL106579/HL/NHLBI NIH HHS/United States
- HL106579/HL/NHLBI NIH HHS/United States
- R01 HL089940/HL/NHLBI NIH HHS/United States
- R01 HL076686-05/HL/NHLBI NIH HHS/United States
- HL89940/HL/NHLBI NIH HHS/United States
- R01 HL076686/HL/NHLBI NIH HHS/United States
- HL076686/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases