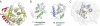Structural and biochemical analysis of nuclease domain of clustered regularly interspaced short palindromic repeat (CRISPR)-associated protein 3 (Cas3) - PubMed (original) (raw)
Structural and biochemical analysis of nuclease domain of clustered regularly interspaced short palindromic repeat (CRISPR)-associated protein 3 (Cas3)
Sabin Mulepati et al. J Biol Chem. 2011.
Abstract
RNA transcribed from clustered regularly interspaced short palindromic repeats (CRISPRs) protects many prokaryotes from invasion by foreign DNA such as viruses, conjugative plasmids, and transposable elements. Cas3 (CRISPR-associated protein 3) is essential for this CRISPR protection and is thought to mediate cleavage of the foreign DNA through its N-terminal histidine-aspartate (HD) domain. We report here the 1.8 Å crystal structure of the HD domain of Cas3 from Thermus thermophilus HB8. Structural and biochemical studies predict that this enzyme binds two metal ions at its active site. We also demonstrate that the single-stranded DNA endonuclease activity of this T. thermophilus domain is activated not by magnesium but by transition metal ions such as manganese and nickel. Structure-guided mutagenesis confirms the importance of the metal-binding residues for the nuclease activity and identifies other active site residues. Overall, these results provide a framework for understanding the role of Cas3 in the CRISPR system.
Figures
FIGURE 1.
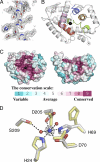
Crystal structure of T. thermophilus Cas3HDdom. A, unbiased Fo − Fc electron density map contoured at 3σ. The residues, which are represented as sticks, were omitted from the map calculation. B, ribbon trace of the Cas3HDdom structure. The HD domain motifs are colored as follows: motif I (red), motif II (green), motif III (orange), motif IV (blue) and motif V (pink). The black sphere represents the cognate metal binding site. The N and C termini are labeled. C, Amino acid sequence conservation scores mapped onto the surface of two orthogonal views of the Cas3HDdom using CONSURF (41). The structure to the left is in the same orientation as shown in B. The conservation scale is drawn below the two views of the structure. The black sphere represents the cognate metal-binding site. D, Cas3HDdom metal ion-binding site. Residues colored by element are in the metal ion-bound configuration. Residues colored yellow are in the protein alone configuration. The Ni2+ (blue) and two water molecules (red) are represented as spheres. Anomalous difference electron density map contoured at 5σ (white mesh) reveals the site of the Ni2+ ion.
FIGURE 2.
Comparison of Cas3HDdom with other HD domains. A, structure of T. thermophilus Cas3HDdom (yellow and red) superimposed on the structure of M. jannaschii Cas3″ (white). The additional helix-strand-strand element in the structure of Cas3HDdom is colored red. The molecules are oriented as shown in B. B, side-by-side view of the structures of T. thermophilus Cas3HDdom (left) and M. jannaschii Cas3″ (right). The helical insertion found in M. jannaschii Cas3″ and its equivalent loop in T. thermophilus Cas3 are colored blue. The helices either side of this region are colored green. C, ribbon trace of the minimal core five α-helices of the HD domain taken from the structure of Cas3HDdom. Residues from the five HD domain motifs are colored as shown in B. A surface representation of Cas3HDdom is shown in the background.
FIGURE 3.
Metal ion-binding sites. The residues that form the metal ion-binding sites of M. jannaschii Cas3″ (A), T. thermophilus Cas3HDdom (B), and a protein of unknown function (PDB 2PQ7) from a Thermotogales species (C). The text color of residue labels indicates the motif that the residue belongs to as shown in Fig. 1_B_. Labeled metal ions are shown as green spheres. His-138 of T. thermophilus Cas3HDdom is modeled in two alternative conformers; for clarity, only one of these conformers is shown.
FIGURE 4.
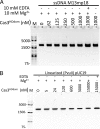
The nuclease activity of Cas3HDdom is not activated by Mg2+. A, activity of Cas3HDdom on ssDNA in the presence of Mg2+. Reaction mixtures containing 10 m
m
Tris-HCl, pH 7.5, 60 m
m
KCl, 10 m
m
MgCl2, 10% glycerol, and 4 n
m
circular single-stranded M13mp18, and the indicated amounts of Cas3HDdom were incubated for 2 h at 37 °C. Reactions with EDTA or no added metal served as controls. All reactions were quenched with 20 m
m
EDTA, and the products were then resolved by a 1% agarose gel and visualized by ethidium bromide staining. B, activity of Cas3HDdom on dsDNA in the presence of Mg2+. Assays were performed as above except that the reaction mixtures contained 4 n
m
pUC19 linearized with PvuII.
FIGURE 5.
The ssDNA endonuclease activity of Cas3HDdom is activated by transition metal ions. A, activity of Cas3HDdom in the presence of difference divalent metal ions. Reaction mixtures containing 10 m
m
Tris-HCl, pH 7.5, 60 m
m
KCl, 10% glycerol, 1 μ
m
Cas3HDdom, and 4 n
m
circular single-stranded M13mp18 DNA, and the indicated amount of each divalent metal ion were incubated for 2 h at 37 °C. Reactions with EDTA, no added metal (NM) or no protein (C) served as controls. All reactions were quenched with 20 m
m
EDTA, and the products were then resolved by a 1% agarose gel and visualized by ethidium bromide staining. B, activity of Cas3HDdom on dsDNA in the presence of different divalent metal ions. Assays were performed as above except that reaction contained 4 n
m
pUC19 linearized with PvuII.
FIGURE 6.
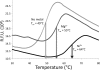
Effects of metal ions on the thermal stability of Cas3HDdom. Thermofluor assays were performed in the absence of metal or in the presence of either 20 m
m
Mg2+ or 100 μ
m
Ni2+. The apparent melting temperatures are indicated by arrows. R.F.U., relative fluorescence units.
FIGURE 7.
Mutational analysis of Cas3HDdom. A, ribbon trace of Cas3HDdom (white). Residues selected for mutation are represented as sticks (red), and the Ni2+ ion as a sphere (blue). B, an SDS-PAGE of the purified mutants, stained with Coomassie Blue. C, ssDNA endonuclease activity of the mutants. Reaction mixtures containing 10 m
m
Tris-HCl, pH 7.5, 60 m
m
KCl, 10% glycerol, 100 n
m
Cas3HDdom, and 60 μ
m
NiSO4, and 4 n
m
circular single-stranded M13mp18 DNA were incubated for 50 min at 37 °C. Reactions with EDTA or no Cas3HDdom (−) served as controls. The reaction products were resolved by a 1% agarose gel and visualized by ethidium bromide staining.
Similar articles
- Molecular insights into DNA interference by CRISPR-associated nuclease-helicase Cas3.
Gong B, Shin M, Sun J, Jung CH, Bolt EL, van der Oost J, Kim JS. Gong B, et al. Proc Natl Acad Sci U S A. 2014 Nov 18;111(46):16359-64. doi: 10.1073/pnas.1410806111. Epub 2014 Nov 3. Proc Natl Acad Sci U S A. 2014. PMID: 25368186 Free PMC article. - Structure and activity of the Cas3 HD nuclease MJ0384, an effector enzyme of the CRISPR interference.
Beloglazova N, Petit P, Flick R, Brown G, Savchenko A, Yakunin AF. Beloglazova N, et al. EMBO J. 2011 Oct 18;30(22):4616-27. doi: 10.1038/emboj.2011.377. EMBO J. 2011. PMID: 22009198 Free PMC article. - Cas3 is a single-stranded DNA nuclease and ATP-dependent helicase in the CRISPR/Cas immune system.
Sinkunas T, Gasiunas G, Fremaux C, Barrangou R, Horvath P, Siksnys V. Sinkunas T, et al. EMBO J. 2011 Apr 6;30(7):1335-42. doi: 10.1038/emboj.2011.41. Epub 2011 Feb 22. EMBO J. 2011. PMID: 21343909 Free PMC article. - Fitting CRISPR-associated Cas3 into the helicase family tree.
Jackson RN, Lavin M, Carter J, Wiedenheft B. Jackson RN, et al. Curr Opin Struct Biol. 2014 Feb;24:106-14. doi: 10.1016/j.sbi.2014.01.001. Epub 2014 Jan 28. Curr Opin Struct Biol. 2014. PMID: 24480304 Free PMC article. Review. - Cas3 Protein-A Review of a Multi-Tasking Machine.
He L, St John James M, Radovcic M, Ivancic-Bace I, Bolt EL. He L, et al. Genes (Basel). 2020 Feb 18;11(2):208. doi: 10.3390/genes11020208. Genes (Basel). 2020. PMID: 32085454 Free PMC article. Review.
Cited by
- An archaeal Cas3 protein facilitates rapid recovery from DNA damage.
Miezner G, Turgeman-Grott I, Zatopek KM, Gardner AF, Reshef L, Choudhary DK, Alstetter M, Allers T, Marchfelder A, Gophna U. Miezner G, et al. Microlife. 2023 Feb 9;4:uqad007. doi: 10.1093/femsml/uqad007. eCollection 2023. Microlife. 2023. PMID: 37223740 Free PMC article. - Anti-CRISPR proteins function through thermodynamic tuning and allosteric regulation of CRISPR RNA-guided surveillance complex.
Patterson A, White A, Waymire E, Fleck S, Golden S, Wilkinson RA, Wiedenheft B, Bothner B. Patterson A, et al. Nucleic Acids Res. 2022 Oct 28;50(19):11243-11254. doi: 10.1093/nar/gkac841. Nucleic Acids Res. 2022. PMID: 36215034 Free PMC article. - Characterization of the self-targeting Type IV CRISPR interference system in Pseudomonas oleovorans.
Guo X, Sanchez-Londono M, Gomes-Filho JV, Hernandez-Tamayo R, Rust S, Immelmann LM, Schäfer P, Wiegel J, Graumann PL, Randau L. Guo X, et al. Nat Microbiol. 2022 Nov;7(11):1870-1878. doi: 10.1038/s41564-022-01229-2. Epub 2022 Sep 29. Nat Microbiol. 2022. PMID: 36175516 - Dynamic mechanisms of CRISPR interference by Escherichia coli CRISPR-Cas3.
Yoshimi K, Takeshita K, Kodera N, Shibumura S, Yamauchi Y, Omatsu M, Umeda K, Kunihiro Y, Yamamoto M, Mashimo T. Yoshimi K, et al. Nat Commun. 2022 Aug 30;13(1):4917. doi: 10.1038/s41467-022-32618-0. Nat Commun. 2022. PMID: 36042215 Free PMC article. - Metal Dependence and Functional Diversity of Type I Cas3 Nucleases.
Sun S, He Z, Jiang P, Baral R, Pandelia ME. Sun S, et al. Biochemistry. 2022 Mar 1;61(5):327-338. doi: 10.1021/acs.biochem.1c00779. Epub 2022 Feb 21. Biochemistry. 2022. PMID: 35184547 Free PMC article.
References
- Mojica F. J., Díez-Villaseñor C., García-Martínez J., Soria E. (2005) J. Mol. Evol. 60, 174–182 - PubMed
- Pourcel C., Salvignol G., Vergnaud G. (2005) Microbiology 151, 653–663 - PubMed
- Bolotin A., Quinquis B., Sorokin A., Ehrlich S. D. (2005) Microbiology 151, 2551–2561 - PubMed
- Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S., Romero D. A., Horvath P. (2007) Science 315, 1709–1712 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM097330/GM/NIGMS NIH HHS/United States
- R01 GM097330-01A1/GM/NIGMS NIH HHS/United States
- T32 CA009110/CA/NCI NIH HHS/United States
- GM097330/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources