Allosteric model of maraviroc binding to CC chemokine receptor 5 (CCR5) - PubMed (original) (raw)
Allosteric model of maraviroc binding to CC chemokine receptor 5 (CCR5)
Javier Garcia-Perez et al. J Biol Chem. 2011.
Abstract
Maraviroc is a nonpeptidic small molecule human immunodeficiency virus type 1 (HIV-1) entry inhibitor that has just entered the therapeutic arsenal for the treatment of patients. We recently demonstrated that maraviroc binding to the HIV-1 coreceptor, CC chemokine receptor 5 (CCR5), prevents it from binding the chemokine CCL3 and the viral envelope glycoprotein gp120 by an allosteric mechanism. However, incomplete knowledge of ligand-binding sites and the lack of CCR5 crystal structures have hampered an in-depth molecular understanding of how the inhibitor works. Here, we addressed these issues by combining site-directed mutagenesis (SDM) with homology modeling and docking. Six crystal structures of G-protein-coupled receptors were compared for their suitability for CCR5 modeling. All CCR5 models had equally good geometry, but that built from the recently reported dimeric structure of the other HIV-1 coreceptor CXCR4 bound to the peptide CVX15 (Protein Data Bank code 3OE0) best agreed with the SDM data and discriminated CCR5 from non-CCR5 binders in a virtual screening approach. SDM and automated docking predicted that maraviroc inserts deeply in CCR5 transmembrane cavity where it can occupy three different binding sites, whereas CCL3 and gp120 lie on distinct yet overlapped regions of the CCR5 extracellular loop 2. Data suggesting that the transmembrane cavity remains accessible for maraviroc in CCL3-bound and gp120-bound CCR5 help explain our previous observation that the inhibitor enhances dissociation of preformed ligand-CCR5 complexes. Finally, we identified residues in the predicted CCR5 dimer interface that are mandatory for gp120 binding, suggesting that receptor dimerization might represent a target for new CCR5 entry inhibitors.
Figures
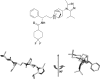
FIGURE 1.
Chemical structure of maraviroc (top) and presumed active three-dimensional structure of maraviroc (bottom).
FIGURE 2.
Snake plot of human CCR5 sequence. Residues tested in this study are highlighted with a black background.
FIGURE 3.
Multiple binding modes for maraviroc into CCR5. All (left) or Glu-283 interacting (right) docked poses of MVC are displayed as thin lines, with CPK color coding of the atomic bonds. The 7TMs of CCR5 are represented by cylinders, as viewed from the extracellular side of the receptor (top) or in the plane of the plasma membrane (bottom). The position of ligands cocrystallized with CXCR4 are indicated in green using ball-and-stick and ribbon representations for the synthetic molecule IT1t and the peptide CVX15, respectively.
FIGURE 4.
Effects of CCR5 single mutations on CCL3 (left), gp120 (middle), and MVC (right) binding to the receptor. As in Fig. 3, the 7TM of CCR5 are represented by cylinders, as viewed from the extracellular side of the receptor (top) or in the plane of the plasma membrane (bottom). The side chains of mutated residues are displayed as capped sticks. Atoms are colored as follows: oxygen in red, nitrogen in blue, sulfur in pale yellow, carbon using a white-yellow-red color scale depending on the strength of radioligand binding inhibition (white denotes unchanged binding upon mutation, and red indicates that binding is lost). No accurate measurements could be reported for residues colored in magenta.
FIGURE 5.
Three-dimensional views of CCR5 model, together with CCL3 crystal structure (left) and gp120 crystal structure (right). The 7TM of CCR5 are represented by cylinders, as viewed from the extracellular side of the receptor (top) or in the plane of the plasma membrane (bottom). The solution structure of CCL3 was extracted from 1B50 PDB entry. It is represented by a green ribbon. Its orientation was chosen based on the electrostatic potentials of the two proteins as follows: the positively charged side of CCL3 faces the negatively charged entry of the membrane receptor. The two proteins were docked to establish H-bonds between the ECL2 hairpin and the CCL3 N-terminal β-strand. Note that the N terminus of the chemokine is truncated (it starts with Asp-5) and that its conformation is not relevant (it bumps into the receptor). The complex between gp120 (green ribbon), CD4 (light blue ribbon), and an antibody (dark blue ribbon) was extracted from 2QAD PDB entry. The antibody includes two sulfotyrosine residues which mimic that of CCR5 N terminus at positions 10 and 14. The orientation was chosen to establish H-bonds between the backbone atoms of CCR5-ECL2 and the tip of gp120V3 loop, according to what was observed between the CXCR4 structure template and the cocrystal peptide CVX15 (yellow ribbon). Note that this orientation is in agreement with an interaction between the gp120 hinge region (between V3 loop and the core protein) and CCR5 sulfotyrosine residues (the CCR5 model here starts at position 17).
Similar articles
- New insights into the mechanisms whereby low molecular weight CCR5 ligands inhibit HIV-1 infection.
Garcia-Perez J, Rueda P, Staropoli I, Kellenberger E, Alcami J, Arenzana-Seisdedos F, Lagane B. Garcia-Perez J, et al. J Biol Chem. 2011 Feb 18;286(7):4978-90. doi: 10.1074/jbc.M110.168955. Epub 2010 Nov 30. J Biol Chem. 2011. PMID: 21118814 Free PMC article. - Structure of the CCR5 chemokine receptor-HIV entry inhibitor maraviroc complex.
Tan Q, Zhu Y, Li J, Chen Z, Han GW, Kufareva I, Li T, Ma L, Fenalti G, Li J, Zhang W, Xie X, Yang H, Jiang H, Cherezov V, Liu H, Stevens RC, Zhao Q, Wu B. Tan Q, et al. Science. 2013 Sep 20;341(6152):1387-90. doi: 10.1126/science.1241475. Epub 2013 Sep 12. Science. 2013. PMID: 24030490 Free PMC article. - A study of the molecular mechanism of binding kinetics and long residence times of human CCR5 receptor small molecule allosteric ligands.
Swinney DC, Beavis P, Chuang KT, Zheng Y, Lee I, Gee P, Deval J, Rotstein DM, Dioszegi M, Ravendran P, Zhang J, Sankuratri S, Kondru R, Vauquelin G. Swinney DC, et al. Br J Pharmacol. 2014 Jul;171(14):3364-75. doi: 10.1111/bph.12683. Br J Pharmacol. 2014. PMID: 24628038 Free PMC article. - Modeling the allosteric modulation of CCR5 function by Maraviroc.
Lagane B, Garcia-Perez J, Kellenberger E. Lagane B, et al. Drug Discov Today Technol. 2013 Summer;10(2):e297-305. doi: 10.1016/j.ddtec.2012.07.011. Drug Discov Today Technol. 2013. PMID: 24050281 Review. - [Viral entry as therapeutic target. Current situation of entry inhibitors].
Arenzana-Seisdedos F. Arenzana-Seisdedos F. Enferm Infecc Microbiol Clin. 2008 Oct;26 Suppl 11:5-11. doi: 10.1016/s0213-005x(08)76557-1. Enferm Infecc Microbiol Clin. 2008. PMID: 19133215 Review. Spanish.
Cited by
- Identification and interaction mechanism of novel small molecule antagonists targeting CC chemokine receptor 1/3/5 for treatment of non-small cell lung cancer.
Wang X, Wang J, Fu Q, Luo J, Shu M, Lin Z. Wang X, et al. Mol Divers. 2024 Nov 22. doi: 10.1007/s11030-024-11025-1. Online ahead of print. Mol Divers. 2024. PMID: 39578293 - CCR5 promotes the migration of pathological CD8+ T cells to the leishmanial lesions.
Amorim Sacramento L, Farias Amorim C, G Lombana C, Beiting D, Novais F, P Carvalho L, M Carvalho E, Scott P. Amorim Sacramento L, et al. PLoS Pathog. 2024 May 6;20(5):e1012211. doi: 10.1371/journal.ppat.1012211. eCollection 2024 May. PLoS Pathog. 2024. PMID: 38709823 Free PMC article. - JAK/STAT signaling pathway affects CCR5 expression in human CD4+ T cells.
Wang L, Yukselten Y, Nuwagaba J, Sutton RE. Wang L, et al. Sci Adv. 2024 Mar 22;10(12):eadl0368. doi: 10.1126/sciadv.adl0368. Epub 2024 Mar 20. Sci Adv. 2024. PMID: 38507500 Free PMC article. - The chemokine receptor CCR5: multi-faceted hook for HIV-1.
Faivre N, Verollet C, Dumas F. Faivre N, et al. Retrovirology. 2024 Jan 23;21(1):2. doi: 10.1186/s12977-024-00634-1. Retrovirology. 2024. PMID: 38263120 Free PMC article. Review. - CCR5 promotes the migration of CD8+ T cells to the leishmanial lesions.
Sacramento LA, Amorim CF, Lombana CG, Beiting D, Novais F, Carvalho LP, Carvalho EM, Scott P. Sacramento LA, et al. bioRxiv [Preprint]. 2023 Oct 13:2023.10.10.561700. doi: 10.1101/2023.10.10.561700. bioRxiv. 2023. PMID: 37873253 Free PMC article. Updated. Preprint.
References
- Blanpain C., Migeotte I., Lee B., Vakili J., Doranz B. J., Govaerts C., Vassart G., Doms R. W., Parmentier M. (1999) Blood 94, 1899–1905 - PubMed
- Alkhatib G., Combadiere C., Broder C. C., Feng Y., Kennedy P. E., Murphy P. M., Berger E. A. (1996) Science 272, 1955–1958 - PubMed
- Arenzana-Seisdedos F., Parmentier M. (2006) Semin. Immunol. 18, 387–403 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources