Proteases involved in cartilage matrix degradation in osteoarthritis - PubMed (original) (raw)
Review
Proteases involved in cartilage matrix degradation in osteoarthritis
Linda Troeberg et al. Biochim Biophys Acta. 2012 Jan.
Abstract
Osteoarthritis is a common joint disease for which there are currently no disease-modifying drugs available. Degradation of the cartilage extracellular matrix is a central feature of the disease and is widely thought to be mediated by proteinases that degrade structural components of the matrix, primarily aggrecan and collagen. Studies on transgenic mice have confirmed the central role of Adamalysin with Thrombospondin Motifs 5 (ADAMTS-5) in aggrecan degradation, and the collagenolytic matrix metalloproteinase MMP-13 in collagen degradation. This review discusses recent advances in current understanding of the mechanisms regulating expression of these key enzymes, as well as reviewing the roles of other proteinases in cartilage destruction. This article is part of a Special Issue entitled: Proteolysis 50 years after the discovery of lysosome.
Copyright © 2011 Elsevier B.V. All rights reserved.
Figures
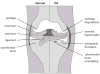
Figure 1. Cartoon representation of normal and osteoarthritic joint
OA is characterised by changes to various tissues within synovial joints. The cartilage matrix is degraded by collagenases and aggrecanases, leading to fibrillation and subsequent loss of the articulating cartilage surface. Synovial fibroblasts undergo hypertrophy and inflammatory cells infiltrate the synovium. Bone remodelling leads to the formation of osteophytes at the cartilage/bone interface and subchondral bone sclerosis.
Figure 2. Schematic representation of domain structure of proteinases involved in OA cartilage destruction
The MMPs, ADAMTSs and ADAMs all contain an N-terminal signal peptide (SP), followed by a pro-domain (pro) and a metalloproteinase catalytic domain. The MMPs then contain a hemopexin ancillary domain [233], while the ADAMTSs contain disintegrin, thrombospondin (TS), cysteine-rich (CysR) and spacer ancillary domains [234]. The ADAMs contain C-terminal disintegrin, CysR, epidermal growth factor-like (EGF-like), transmembrane (TM) and cytoplasmic (Cyt) domains [235]. The serine proteinases involved in OA cartilage destruction are more structurally diverse than the metalloproteinases. uPA consists of an N-terminal SP, followed by an EGF-like domain, a Kringle domain and a C-terminal catalytic domain [236]. Matriptase 1 is a type II transmembrane protein, with an N-terminal cytoplasmic domain followed by a TM region [190]. This is followed by a sea urchin sperm protein/enteropeptidase/agrin (SEA) domain, 2 complement C1r/C1s, Uegf, Bmp1 (CUB) domains, 4 low-density lipoprotein receptor-related protein (LRP) domains and a C-terminal catalytic domain [190]. APC consists of a γ-carboxyglutamate (Gla) domain, followed by 2 EGF-domains and a trypsin-like serine proteinase domain [237]. Proprotein convertases such as furin are subtilisin-like serine proteinases consisting of an N-terminal SP and pro-domain, followed by a serine proteinase catalytic domain, a conserved regulatory P domain and a CysR domain [238]. Some of the proprotein convertases (e. g. furin) are type I transmembrane proteins and contain a C-terminal TM and cytoplasmic domain, while others (e. g. PACE4) lack these domains and are soluble [238]. HtrA1 consists of an N-terminal SP, followed by an insulin growth factor binding protein (IGFBP) domain, Kazal proteinase inhibitor (KI) domain, a trypsin-like serine proteinase domain and a C-terminal PDZ domain [199]. The cathepsins have comparatively simple structures, consisting of an SP, pro-domain and catalytic domain with no additional ancillary domains [239]. Classical calpains such as m-calpain consist of 4 domains, with domain I (D1) and domain II (DII) forming the catalytic domain, and domains III and IV (DIII and DIV) regulating catalytic activity and stability [240]. Domain III is C2-like, and domain IV contains 5 EF-hand repeats. Caspases have all contain a large and a small catalytic subunit. N-terminal to this is either an N-terminal caspase recruitment domain (CARD) domain (e. g. in caspase 1, 2, 4 and 5) or an N-terminal death effector domain (DED) (e. g. in caspase 8, 10) [241].
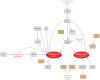
Figure 3. Factors regulating expression and activity of collagenases and aggrecanases in OA
The expression and activity of collagenases (e. g. MMP-13 and MMP-1) and aggrecanases (e. g. ADAMTS-4 and ADAMTS-5) can be stimulated (orange boxes) or inhibited (grey boxes) by a number of inter-related mechanisms. Expression of the central transcription factor RUNX2 is increased by mechanical and pro-inflammatory stimuli, which act via HIF-2α, HDACs and Indian hedgehog. Expression of collagenolytic MMPs can also be increased in response to FGF-2 and DDR-2 signalling, and collagenase activity increased by matriptase activation of proMMP-1 and proMMP-3 zymogens and PAR-2 signalling. These catabolic stimuli can be counteracted by a variety of chondroprotective signals. For example, MMP13 expression can be reduced by the mechano-sensitive transcription factor CITED, and ADAMTS5 expression can be reduced by miR-140 and the mechano-responsive growth factor FGF-2.
Similar articles
- [Destruction of the articular cartilage in osteoarthritis].
Okubo M, Okada Y. Okubo M, et al. Clin Calcium. 2013 Dec;23(12):1705-13. Clin Calcium. 2013. PMID: 24292524 Review. Japanese. - MMP proteolysis of the human extracellular matrix protein aggrecan is mainly a process of normal turnover.
Struglics A, Hansson M. Struglics A, et al. Biochem J. 2012 Sep 1;446(2):213-23. doi: 10.1042/BJ20120274. Biochem J. 2012. PMID: 22670872 - [Progress of research in osteoarthritis. Metalloproteinases in osteoarthritis].
Okada A, Okada Y. Okada A, et al. Clin Calcium. 2009 Nov;19(11):1593-601. Clin Calcium. 2009. PMID: 19880991 Review. Japanese. - The effect of protease inhibitors on the induction of osteoarthritis-related biomarkers in bovine full-depth cartilage explants.
He Y, Zheng Q, Jiang M, Sun S, Christiansen TG, Kassem M, Karsdal MA, Bay-Jensen AC. He Y, et al. PLoS One. 2015 Apr 24;10(4):e0122700. doi: 10.1371/journal.pone.0122700. eCollection 2015. PLoS One. 2015. PMID: 25909781 Free PMC article. - Matrix metalloproteinases: role in arthritis.
Burrage PS, Mix KS, Brinckerhoff CE. Burrage PS, et al. Front Biosci. 2006 Jan 1;11:529-43. doi: 10.2741/1817. Front Biosci. 2006. PMID: 16146751 Review.
Cited by
- MicroRNA-613 alleviates IL-1β-induced injury in chondrogenic CHON-001 cells by targeting fibronectin 1.
Xiao P, Zhu X, Sun J, Zhang Y, Qiu W, Li J, Wu X. Xiao P, et al. Am J Transl Res. 2020 Sep 15;12(9):5308-5319. eCollection 2020. Am J Transl Res. 2020. PMID: 33042421 Free PMC article. - Novel Insights into Osteoarthritis Joint Pathology from Studies in Mice.
Moon PM, Beier F. Moon PM, et al. Curr Rheumatol Rep. 2015 Aug;17(8):50. doi: 10.1007/s11926-015-0524-1. Curr Rheumatol Rep. 2015. PMID: 26113010 Review. - Matrix Metalloproteinases in Non-Neoplastic Disorders.
Tokito A, Jougasaki M. Tokito A, et al. Int J Mol Sci. 2016 Jul 21;17(7):1178. doi: 10.3390/ijms17071178. Int J Mol Sci. 2016. PMID: 27455234 Free PMC article. Review. - Osteoarthritis: a disease of the joint as an organ.
Loeser RF, Goldring SR, Scanzello CR, Goldring MB. Loeser RF, et al. Arthritis Rheum. 2012 Jun;64(6):1697-707. doi: 10.1002/art.34453. Epub 2012 Mar 5. Arthritis Rheum. 2012. PMID: 22392533 Free PMC article. Review. No abstract available. - Transcription Factors in Cartilage Homeostasis and Osteoarthritis.
Neefjes M, van Caam APM, van der Kraan PM. Neefjes M, et al. Biology (Basel). 2020 Sep 14;9(9):290. doi: 10.3390/biology9090290. Biology (Basel). 2020. PMID: 32937960 Free PMC article. Review.
References
- Wieland HA, Michaelis M, Kirschbaum BJ, Rudolphi KA. Osteoarthritis - an untreatable disease? Nature Reviews Drug discovery. 2005;4:331–344. - PubMed
- Goldring MB, Goldring SR. Articular cartilage and subchondral bone in the pathogenesis of osteoarthritis. Ann N Y Acad Sci. 2010;1192:230–237. - PubMed
- Heinegård D, Saxne T. The role of the cartilage matrix in osteoarthritis. Nat Rev Rheumatol. 2011;7:50–56. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AR040994/AR/NIAMS NIH HHS/United States
- 19466/ARC_/Arthritis Research UK/United Kingdom
- R01 AR040994-18/AR/NIAMS NIH HHS/United States
- AR40994/AR/NIAMS NIH HHS/United States
- 19466/VAC_/Versus Arthritis/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases