Autophagosome precursor maturation requires homotypic fusion - PubMed (original) (raw)
Autophagosome precursor maturation requires homotypic fusion
Kevin Moreau et al. Cell. 2011.
Abstract
Autophagy is a catabolic process in which lysosomes degrade intracytoplasmic contents transported in double-membraned autophagosomes. Autophagosomes are formed by the elongation and fusion of phagophores, which can be derived from preautophagosomal structures coming from the plasma membrane and other sites like the endoplasmic reticulum and mitochondria. The mechanisms by which preautophagosomal structures elongate their membranes and mature toward fully formed autophagosomes still remain unknown. Here, we show that the maturation of the early Atg16L1 precursors requires homotypic fusion, which is essential for subsequent autophagosome formation. Atg16L1 precursor homotypic fusion depends on the SNARE protein VAMP7 together with partner SNAREs. Atg16L1 precursor homotypic fusion is a critical event in the early phases of autophagy that couples membrane acquisition and autophagosome biogenesis, as this step regulates the size of the vesicles, which in turn appears to influence their subsequent maturation into LC3-positive autophagosomes.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures
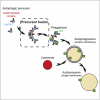
Graphical abstract
Figure 1
VAMP7 Colocalizes with Atg16L1 and Atg5 (A) Colocalization between LC3-CFP, Atg16L1-GFP, Atg16L1-Flag, Atg5-GFP, VAMP7-mRFP, endogenous VAMP7, and stably expressed VAMP7-HA. Cells were transfected for 20 hr. Confocal colocalization is indicated. At least 20 cells were analyzed in each condition. Error bars = standard deviation (SD). Scale bar, 5 μm. (B) HeLa cells stably expressing VAMP7-HA were transfected for 20 hr with Atg16L1-GFP. The cells were processed for double immunogold labeling with anti-HA antibodies (15 nm gold particles) and anti-GFP antibodies (10 nm gold particles) for electron microscopy. Colocalization between Atg16L1 structures with VAMP7-HA gold particles is shown. n = 17 cells. Scale bar, 500 nm. Error bars = standard error of the mean (SEM). See also Figure S1.
Figure 2
VAMP7 Regulates Autophagic Activity (A) HeLa cells transfected with control or VAMP7 siRNA for 5 days were treated during the last 16 hr with bafilomycin A1 (Baf A1) or DMSO. Means ± SD of the percentage of LC3-II/tubulin ratio from three independent experiments are shown. LE (light exposure of film), HE (longer exposure of film). (B) HeLa cells transfected with control or VAMP7 siRNA for 5 days were starved and treated during the last 16 hr with bafilomycin A1 (Baf A1) or DMSO. Means ± SD of LC3-II/actin ratio from three independent experiments. (C) HeLa cells stably expressing LC3-GFP-mRFP were transfected with control or VAMP7 siRNA for 5 days and treated during the last 16 hr with bafilomycin A1 (Baf A1) or DMSO. Cells were fixed and subjected to automated counting of LC3 vesicles. Representative confocal pictures are shown. Scale bar, 5 μm. (D) Quantification of total number of vesicles/cell, autophagosomes/cell (AV/cell; LC3 GFP+/mRFP+), and autolysosomes/cell (AL/cell; LC3 GFP−/mRFP+) from (C) is shown. At least 2000 cells were counted per experiment; values are mean ± SEM of one representative experiment of three independent experiments performed. (E) HeLa cells transfected with control, VAMP7, Ulk1, or Sec22B siRNA for 5 days were immunolabeled for endogenous LC3 (red) and Atg16L1 (green). Representative confocal pictures and quantification of Atg16L1 vesicles are shown. At least 50 cells were counted per experiment; values are mean ± SD of the number of Atg16L1 vesicles per cell obtained from two independent experiments. Colocalization efficiency between Atg16L1 and LC3 is also shown. At least 50 cells were counted per experiment and the data represent the percentage of Atg16L1 vesicles containing LC3 ± SD obtained from two independent experiments. Scale bar, 5 μm. NS—not significant. See also Figure S2.
Figure 3
VAMP7 Mediates Homotypic Fusion of Atg16L1 Vesicle (A) HeLa cells transfected with control, VAMP7, syntaxin 7 (Stx7), syntaxin 8 (Stx8), or Vti1b siRNA for 4 days were transfected for 20 hr with Atg16L1-GFP. Representative images from 10 min movies are shown. Scale bars, 5 μm. (B) Quantification of the number of fusion events in 10 min (fusion and kiss and run) from the movies obtained in (A) is shown. Data represent the mean ± SEM of the number of fusion events per cell in 10 min. n = 5 cells for control, n = 6 cells for VAMP7 knockdown, n = 9 cells for Stx7 knockdown, n = 5 cells for Stx8 knockdown, and n = 6 cells for Vti1b knockdown. (C) The velocity of the Atg16L1-GFP vesicles from the experiment described in (A), obtained using ImageJ manual tracking plugin. Data represent the mean ± SEM of the velocity of 10 Atg16L1 vesicles. NS—not significant. (D) HeLa cells transfected with Atg16L1-GFP were subjected to live-cell imaging. Representative images (in inverted gray mode) from 10 min movies are shown (Untreated). Then, N-ethylmaleimide (NEM; 100 μM) was added to the medium and the acquisition started again for 10 min. Representative images from 10 min movies are shown (NEM). Arrows indicate fusion between two Atg16L1-GFP vesicles in untreated condition, whereas arrows indicate no fusion event in NEM-treated condition. Scale bars, 5 μm. (E) Numbers of fusion events in 10 min (fusion and kiss and run) from the movies obtained in (D). Data represent the mean ± SD of the number of fusion events per cell in 10 min. n = 5 cells for untreated cells, n = 5 cells for NEM-treated cells. (F) In vitro fusion assay of Atg16L1 vesicles. Postnuclear supernatants (PNS) of cells expressing Atg16L1-GFP were mixed with PNS from cells expressing either Atg16L1-Flag or Atg16L1-mStrawberry as indicated for 1 hr, in presence or absence of ATP and NEM. Representative confocal pictures are shown. The data represent the mean ± SD of the percentage of Atg16L1-Flag or Atg16L1-mStrawberry vesicles fused (colocalized) with Atg16L1-GFP. n = 202 vesicles for untreated condition, n = 170 vesicles for NEM-treated condition, n = 131 vesicles for ATP-untreated condition obtained from two independent experiments when the Atg16L1-Flag was used, n = 118 vesicles for untreated condition, n = 73 vesicles for NEM-treated condition, n = 31 vesicles for ATP-untreated condition obtained from 1 experiment when Atg16L1-mStrawberry was used. (G) After the in vitro fusion reaction of Atg16L1 vesicles, as described in (F), the vesicles were stained for endogenous LC3. A representative confocal picture is shown where two fused Atg16L1 vesicles (yellow dot) contain LC3 whereas one fused vesicle and one unfused vesicle (green) do not contain LC3. Graph represents the percentage of Atg16L1 vesicles containing LC3 depending on if they are fused (yellow) or unfused (green). Data are the mean ± SD. n = 66 vesicles obtained from two independent experiments. See also Figure S3 and Movie S1, Movie S2, Movie S3, Movie S4, Movie S5, Movie S6, Movie S7, and Movie S8.
Figure 4
VAMP7 Regulates Atg16L1 Vesicle Size (A) HeLa cells were transfected for 20 hr with LC3-CFP (red) and Atg16L1-GFP (green). Confocal colocalization between LC3-CFP and Atg16L1-GFP is shown in white on the image. Yellow arrows show colocalization between LC3 and Atg16L1; white arrows indicate Atg16L1 vesicles with no LC3. Scale bar, 5 μm. (B) Size distributions of total Atg16L1 vesicles and Atg16L1+/LC3+ vesicles from images taken in (A) using ImageJ. n = 5 cells from two independent experiments. (C) HeLa cells transfected with control or VAMP7 siRNA for 4 days were transfected for 20 hr with Atg16L1-GFP. Representative confocal images are shown. Scale bar = 5 μm. (D) Atg16L1-GFP vesicle sizes from (C). Mean ± SEM of the average size of Atg16L1-GFP vesicles (A.U.: arbitrary unit); three independent experiments; at least 2000 cells were automatically analyzed. (E) Size distribution of Atg16L1-GFP vesicles from images taken in (C) using ImageJ. n = 4 cells for control and VAMP7 knockdown; two independent experiments. (F) HeLa cells transfected with control or VAMP7 siRNA for 5 days were starved for 4 hr. Size distribution of endogenous Atg16L1 vesicles is shown. n = 20 cells for control and VAMP7 knockdown; two independent experiments. (G) Representative images from the experiment described in (F). Arrows indicate endogenous Atg16L1 vesicles. Scale bar, 5 μm. (H) HeLa cells transfected with control or VAMP7 siRNA for 4 days were transfected for 20 hr with Atg16L1-GFP. The cells were processed for immunogold labeling with anti-GFP antibodies (15 nm gold particles) for electron microscopy. We observed Atg16L1-positive tubular structures in control cells, but not in VAMP7 knockdown cells (right-hand micrographs). Data represent mean ± SEM of the area of Atg16L1-GFP “cluster” (nm2) and the average diameter of individual Atg16L1-GFP vesicles (nm). n = 13 clusters for control. n = 19 clusters for VAMP7KD. n = 100 single vesicles for control. n = 100 single vesicles for VAMP7KD. See also Figure S4.
Figure 5
N-ethylmaleimide Treatment Decreases Atg16L1 Precursor Maturation (A) HeLa cells transiently transfected for 20 hr with Atg16L1-Flag and Atg5-GFP were treated as indicated with N-ethylmaleimide (NEM; 100 μM) for 10 min. Representative confocal pictures are shown. Scale bars, 5 μm. (B) Size distribution of Atg16L1-Flag vesicles from (A). n > 40 cells for untreated and NEM-treated cells; three independent experiments. (C) Size distribution of Atg5-GFP vesicles from (A). n > 20 cells for untreated and NEM-treated cells from three independent experiments. (D) HeLa cells were treated as indicated with N-ethylmaleimide (NEM; 100 μM) for 10 min. Cells were immunolabeled for endogenous Atg16L1 and LC3 and subjected to confocal analysis. Scale bars, 5 μm. (E) Colocalization efficiency between Atg16L1 and LC3 from (D). At least 50 cells were counted per experiment; data are the mean ± SD of the percentage of Atg16L1 vesicles containing LC3; three independent experiments. (F) HeLa cells were treated as indicated with N-ethylmaleimide (NEM; 100 μM) for 10 min. Cells were immunolabeled for endogenous Atg16L1 and Atg12 and subjected to confocal analysis. Scale bars, 5 μm. At least 50 cells were counted per experiment and the data are mean ± SD of the number of Atg16L1 or Atg12 vesicles per cell; three independent experiments. See also Figure S5.
Figure 6
Hrb and the Longin Domain of VAMP7 Are Involved in Autophagosome Formation (A) HeLa cells transfected with control or Hrb siRNA for 5 days were lysed and subjected to western blotting with the indicated antibodies. (B) HeLa cells transfected with control or Hrb siRNA for 5 days were treated during the last 16 hr with bafilomycin A1 (Baf A1) or DMSO. Cells were lysed and subjected to western blotting with the indicated antibodies. (C) LC3-II/actin ratio from (B). Means ± SD of LC3-II/actin ratio; three independent experiments. (D) HeLa cells transfected with control or Hrb siRNA for 5 days were subjected to Alexa 488-labeled transferrin internalization assay; the data represent the mean ± SD of the transferrin fluorescence intensity; n = 2 independent experiments in triplicate. (E) Sizes of Atg16L1-GFP vesicles upon Hrb knockdown. The data represent the mean ± SEM of the average size of Atg16L1-GFP vesicles (A.U.: arbitrary unit); two independent experiments; at least 2000 cells were automatically analyzed. Representative confocal pictures are shown. Scale bars, 5 μm. (F) HeLa cells transfected with control or Hrb siRNA for 5 days were fixed and subjected to immunolabeling for endogenous LC3 (red) and Atg16L1 (green). Representative confocal pictures and Atg16L1 vesicle analyses are shown. At least 50 cells were counted per experiment; values are mean ± SD of the number of Atg16L1 vesicles per cell; two independent experiments. Colocalization efficiency between Atg16L1 and LC3. At least 50 cells were counted per experiment. Data are mean ± SD of the percentage of Atg16L1 vesicles containing LC3; two independent experiments. Scale bar, 5 μm. NS—not significant. (G) HeLa cells stably expressing VAMP7-HA (VAMP7 WT) or a truncated form of VAMP7-HA without the longin domain (VAMP7 ΔLD) were treated during 16 hr with bafilomycin A1 (Baf A1) or DMSO (vehicle). Cells were subjected to western blotting with the indicated antibodies. (H) Quantification of LC3-II/actin ratio from (G). Mean ± SD of the percentage of LC3-II/actin ratio; three independent experiments. (I) VAMP7 WT or VAMP7 ΔLD cells were transfected for 20 hr with Atg16L1-GFP. Cells were subjected to automated counting of Atg16L1 vesicles. Quantification of Atg16L1-GFP vesicles is shown. At least 2000 cells were counted per experiment and the values are mean ± SEM of one representative experiment of three independent experiments performed. (J) Sizes of Atg16L1-GFP vesicles in VAMP7 WT or VAMP7 ΔLD cells from (I). Data represent the mean ± SEM of the average size of Atg16L1-GFP vesicles (A.U.: arbitrary unit); three independent experiments; at least 2000 cells were automatically analyzed.
Figure 7
Autophagy Stimulation Induces Atg16L1 Vesicle Homotypic Fusion (A) HeLa cells transiently transfected for 20 hr with Atg16L1-GFP were treated as indicated with rapamycin (1 μg/ml) for 1 hr or starved for 1 hr. Representative confocal pictures are shown. Scale bars, 5 μm. (B) Quantification of the number of Atg16L1-GFP vesicles per cell using ImageJ from the pictures obtained in (A). Mean ± SD of the number of Atg16L1-GFP vesicle per cell; three independent experiments; n = 20 cells per experiment. (C) HeLa cells transiently transfected for 20 hr with Atg16L1-GFP were treated as indicated with rapamycin (1 μg/ml) for 1 hr or starved for 1 hr and subjected to live-cell imaging. Data are mean ± SD of the number of fusion events per cell in 10 min from two independent experiments. n = 5 cells for untreated condition, n = 7 cells for starvation condition, and n = 5 cells for rapamycin condition. (D) The velocity of the Atg16L1-GFP vesicles from (C) using ImageJ manual tracking plugin. Means ± SEM of the velocity of ten Atg16L1 vesicles. (E) Model of Atg16L1 precursor maturation. Atg16L1 precursors (Atg16L1+/LC3−) formed, in this example, from the plasma membrane undergo homotypic fusion via a VAMP7-containing SNAREs complex. Atg16L1 precursor homotypic fusion leads to LC3 acquisition and therefore maturation toward phagophores (Atg16L1+/LC3+) and fully formed autophagosomes (Atg16L1−/LC3+). See Movie S9 and Movie S10.
Figure S1
Colocalization Analysis between VAMP7, Syntaxin 7, Syntaxin 8, Vti1b, SNAP23, and Atg16L1, Related to Figure 1 (A) Colocalization between endogenous Atg16L1 and stably expressed VAMP7-HA. HeLa cells stably expressing VAMP7-HA were fixed and subjected to confocal analysis. Colocalization efficiency between endogenous Atg16L1 and VAMP7-HA is shown. n = 20 cells. Error bars throughout this figure are SD. Scale bar, 5 μm. (B) Colocalization between LC3-CFP, Atg16L1-GFP and endogenous syntaxin 7 (Stx7), syntaxin 8 (Stx8), Vti1b and SNAP23. HeLa cells were transiently transfected for 20 hr with LC3-CFP and Atg16L1-GFP. Cells were fixed and subjected to confocal analysis after an immunostaining of Stx7, Stx8, Vti1b, or SNAP23. Colocalization efficiency is shown. n ≥ 20 cells in each condition. Scale bars, 5 μm. (C) HeLa cells transfected for 20 hr with Atg16L1-GFP were processed for double immunogold labeling with anti-HRP antibodies (15 nm gold particles; cholera toxin, used as an endocytic tracer) and anti-GFP antibodies (10 nm gold particles) for electron microscopy or were processed for immunofluorescence analysis after an immunostaining for endogenous Atg12. Colocalization between Atg16L1-GFP and the cholera toxin on vesicular structures is shown on the electronic microscopy picture. Colocalization between Atg16L1-GFP and endogenous Atg12 is shown on confocal pictures. Scale bar, 5 μm.
Figure S2
VAMP7, Syntaxin 7, Syntaxin 8, and Vti1b Regulate Autophagic Activity, Related to Figure 2 (A) HeLa cells transfected with two rounds of control, VAMP7, syntaxin 7 (Stx7), syntaxin 8 (Stx8), Vti1b, or Atg16L1 siRNA for 5 days were lysed and subjected to western blotting with the indicated antibodies. For Atg16L1 knockdown, the cells were starved when indicated using HBBS medium treatment for 8 hr and treated during the last 4 hr with bafilomycin A1 (Baf A1) or DMSO (vehicle). Quantification of LC3-II/actin ratio is shown for Atg16L1 knockdown. The data represent the mean ± SD of the percentage of LC3-II/actin ratio obtained from three independent experiments. (B–D) HeLa cells transfected with two rounds of control, Stx7, Stx8, or Vti1b siRNA for 5 days were treated during the last 16 hr with bafilomycin A1 (Baf A1) or DMSO (vehicle). Cells were lysed and subjected to western blotting with the indicated antibodies. Quantification of LC3-II/actin ratio is shown. The data represent the mean ± SD of the percentage of LC3-II/actin ratio obtained from three independent experiments. (E) HeLa cells stably expressing LC3-GFP-mRFP were treated as indicated during the last 16 hr with bafilomycin A1 (Baf A1), fixed and subjected to a Cellomics ArrayScan VTI HCS Reader analysis. Quantification of the size of autophagic vesicles is shown. The data represent the mean ± SEM of the size of autophagic vesicles (A.U; arbitrary unit) of 1 experiment representative of three independent experiments performed where minimums of 2000 cells were automatically analyzed. (F) HeLa cells stably expressing LC3-GFP-mRFP were transfected with two rounds of control or VAMP7 siRNA for 5 days and treated during the last 16 hr with bafilomycin A1 (Baf A1) or DMSO. Cells were starved for 4 hr, fixed, and subjected to automatic counting of LC3 vesicles. Quantification of total number of vesicles/cell, autophagosomes/cell (AV/cell) and autolysosomes/cell (AL/cell) is shown. At least 2000 cells were counted per experiment and the values are mean ± SEM of one representative experiment of three independent experiments performed. (G) HeLa cells transfected with two rounds of control, VAMP7, Stx7, Stx8, Sec22B, or Vti1b siRNA for 5 days were subjected to Alexa 488-labeled transferrin internalisation assay and the amount of fluorescent-transferrin internalised under different knockdown conditions was measured by FACS and presented in the graph. NS—not significant. (H) HeLa cells transfected with two rounds of control, VAMP7 or Stx7 siRNA for 5 days were lysed and subjected to western blotting with the indicated antibodies. Quantification of p62/actin ratio is shown. The data represent the mean ± SD of the percentage of p62/actin ratio obtained from three independent experiments. (I) HeLa cells transfected with two rounds of control, Stx7, Stx8, or Vti1b siRNA for 5 days were fixed and subjected to immunolabeling for endogenous LC3 and Atg16L1. At least 50 cells were counted per experiment and the values are mean ± SD of the number of Atg16L1 vesicles per cell obtained from two independent experiments. The colocalization efficiency between Atg16L1 and LC3 is also shown. At least 50 cells were counted per experiment and the data represent the percentage of Atg16L1 vesicles containing LC3 obtained from two independent experiments. Scale bar, 5 μm. (J) DFCP1-GFP cells transfected with two rounds of control or VAMP7 siRNA for 5 days were fixed and subjected to confocal analysis for DFCP1 dots counting. At least 50 cells were counted per experiment and the data are mean ± SD of the number of DFCP1 dots per cell (in percentage, relative to control) obtained from two independent experiments.
Figure S3
Homotypic Fusion of Atg16L1 Vesicles Analysis, Related to Figure 3 (A) HeLa cells transfected for 20 hr with Atg16L1-GFP and LC3-mRFP were subjected to live cell imaging. Representative images from 5 min movies are shown. Note that two Atg16L1-GFP vesicles without LC3 fused together. Arrows show Atg16L1-GFP vesicles involved in fusion event. Scale bar, 5 μm. (B) Time course of the in vitro fusion assay of Atg16L1 vesicles. Postnuclear supernatants (PNS) of cells expressing Atg16L1-GFP were mixed with PNS from cells expressing Atg16L1-Flag for 10 min, 30 min, or 50 min. The reaction were put on coverslips and analyzed by confocal microscopy after an immunostaining for Flag. The data represent the mean ± SD of the percentage of Atg16L1-GFP vesicles fused (colocalized) with Atg16L1-Flag. n = 30 vesicles for 0 min, n = 33 vesicles for 10 min, n = 79 vesicles for 30 min, n = 93 vesicles for 50 min obtained from two independent experiments. (C) Immunostaining of endogenous LC3 in the in vitro fusion assay of Atg16L1 vesicles without ATP. After the in vitro fusion reaction without ATP, the Atg16L1 vesicles were stained for endogenous LC3 and analyzed by confocal microscopy. A representative confocal picture is shown where Atg16L1 vesicles do not contain LC3 (arrows) whereas one Atg16L1 vesicle does contain LC3 (arrowhead). The graph represents the percentage of Atg16L1 vesicles with LC3. The data are the mean ± SD n = 60 vesicles obtained from two independent experiments. (D) Postnuclear supernatants (PNS) of cells expressing Atg16L1-GFP were mixed with PNS from cells expressing either Atg16L1-Flag for 1 hr, in absence of ATP or presence of NEM. The reaction was put on coverslips and analyzed by confocal microscopy. On the left, the data represent the average size ± SD of Atg16L1-GFP vesicles (arbitrary unit, A.U). n = 163 vesicles for untreated condition, n = 65 vesicles for NEM-treated condition, n = 159 vesicles for ATP-untreated condition obtained from two independent experiments. On the right, the data represent the average size ± SD of unfused Atg16L1-GFP vesicles with Atg16L1-Flag (only green) or fused Atg16L1-GFP vesicles with Atg16L1-Flag (yellow). n = 25 vesicles for unfused vesicles, n = 25 vesicles for fused vesicles. See also Movie S1.
Figure S4
VAMP7, Syntaxin 7, Syntaxin 8, and Vti1b Regulate Atg16L1 and LC3 Vesicle Size, Related to Figure 4 (A) HeLa cells transfected with two rounds of control siRNA for 4 days were transfected for 20 hr with Atg16L1-GFP and subjected to live cell imaging. Representative images from 10 min movies are shown. Scale bar, 5 μm. (B) The graph represents the size of the vesicles, using ImageJ, identified as dot 1 and dot 2 in the images shown in top panel in (A). (C and D) HeLa cells transfected with two rounds of control, VAMP7, Syntaxin 7 (Stx7), syntaxin 8 (Stx8), or Vti1b siRNA for 4 days were transfected for 20 hr with Atg16L1-GFP. Cells were fixed and subjected to confocal analysis. Representative images are shown. Scale bar = 5 μm. Quantification of the size of Atg16L1-GFP vesicles is presented. The data represent the mean ± SEM of the average size of Atg16L1-GFP vesicles (A.U; abitrary unit) from three independent experiments (2 for Vti1b knockdown) where a minimum of 2000 cells were automatically analyzed using a Cellomics ArrayScan VTI HCS Reader. (E) HeLa cells transfected with two rounds of control, VAMP7 smartpool (sp), deconvoluted VAMP7 (Oligo 1 to Oligo 4) or Sec22B siRNA for 4 days were transfected for 20 hr with Atg16L1-GFP. Cells were fixed and subjected to a Cellomics ArrayScan VTI HCS Reader analysis. Quantification of the size of Atg16L1-GFP vesicles is presented. The data represent the mean ± SEM of the average size of Atg16L1-GFP vesicles (A.U) of one representative experiment from three independent experiments performed where a minimum of 2000 cells were automatically analyzed. (F) HeLa cells transfected with two rounds of control or clathrin siRNA for 4 days were transfected for 20 hr with Atg16L1-GFP. Cells were fixed and subjected to a Cellomics ArrayScan VTI HCS Reader analysis. Quantification of the size of Atg16L1-GFP vesicles is presented. The data represent the mean ± SEM of the average size of Atg16L1-GFP vesicles (A.U) from three independent experiments where minimums of 2000 cells were automatically analyzed. (G) Quantification of the number of Atg16L1-GFP vesicles/cell from the experiment described in (F) is presented. The data represent the mean ± SEM of the number of Atg16L1-GFP vesicles/cell (A.U) from three independent experiments where minimums of 2000 cells were automatically analyzed. (H) HeLa cells stably expressing LC3-GFP-mRFP were transfected with two rounds of control or VAMP7 siRNA for 5 days and either left untreated or starved for 4 hr. Cells were treated as indicated during the last 16 hr with bafilomycin A1 (Baf A1), fixed, and subjected to a Cellomics ArrayScan VTI HCS Reader analysis. Quantification of the size of autophagic vesicles is shown. The data represent the mean ± SEM of the size of autophagic vesicles (A.U; arbitrary unit) of one experiment representative of three independent experiments performed where minimums of 2000 cells were automatically analyzed. (I and J) Graph distribution of the size of autophagic vesicles from Baf A1-untreated cells from images taken in (H) using ImageJ. n = 20 cells for untreated condition and n = 10 cells for starvation condition. (K) HeLa cells stably expressing LC3-GFP-mRFP were transfected with two rounds of control or VAMP7 siRNA for 5 days and processed for immunogold labeling with anti-GFP antibodies (15 nm gold particles) for electron microscopy. Representative images are shown. Scale bar = 300 nm. The data represent the perimeter ± SD of LC3-GFP vesicles (nm). n = 13 vesicles for control. n = 19 vesicles for VAMP7KD. (L) HeLa cells stably expressing LC3-GFP-mRFP were transfected with two rounds of control or Syntaxin 7 (Stx7) siRNA for 5 days. Cells were treated as indicated during the last 16 hr with bafilomycin A1 (Baf A1), fixed, and subjected to a Cellomics ArrayScan VTI HCS Reader analysis. Cells were starved where indicated. Quantification of the size of autophagic vesicles is shown. The data represent the mean ± SEM of the size of autophagic vesicles (A.U) of one experiment representative of three independent experiments performed where minimums of 2000 cells were automatically analyzed. (M) HeLa cells stably expressing LC3-GFP-mRFP were transfected with two rounds of control or clathrin siRNA for 5 days. Cells were treated as indicated during the last 16 hr with bafilomycin A1 (Baf A1), fixed, and subjected to a Cellomics ArrayScan VTI HCS Reader analysis. Quantification of the size of autophagic vesicles is shown. The data represent the mean ± SEM of the size of autophagic vesicles (A.U; arbitrary unit) of one experiment representative of three independent experiments performed where minimums of 2000 cells were automatically analyzed. NS—not significant.
Figure S5
NEM and Tetanus Toxin Regulate Atg16L1 Vesicle Size, Related to Figure 5 (A) HeLa cells were transfected for 20 hr with Atg16L1-GFP and either wild-type NSF (NSF WT) or a dominant-negative mutant of NSF (NSF DN). Cells were fixed and subjected to confocal analysis. Representative images are shown. Scale bar = 5 μm. Graph distribution of the size of Atg16L1-GFP vesicles from confocal pictures using ImageJ is shown. n = 5 cells for NSF WT and NSF DN from two independent experiments. (B) HeLa cells stably expressing LC3-GFP-mRFP were treated with N-ethylmaleimide (NEM; 100 μM) for 10 min as indicated. Cells were fixed and subjected to a Cellomics ArrayScan VTI HCS Reader analysis. Representative confocal pictures and the quantification of the size of autophagic vesicles are shown. The data represent the mean ± SEM of the size of autophagic vesicles (A.U; arbitrary unit) of one experiment representative of three independent experiments performed where at least of 2000 cells were automatically analyzed. Scale bar, 5 μm. (C) HeLa cells were transfected for 20 hr with Atg16L1-GFP and either Tetanus neurotoxin-mCherry (TeNT-mCherry) or an empty vector. Cells were fixed and subjected to confocal analysis. Representative images are shown. Scale bar = 5 μm. Graph distribution of the size of Atg16L1-GFP vesicles from confocal pictures using ImageJ is shown. n = 3 cells for TeNT-mCherry and empty vector from two independent experiments.
Similar articles
- Autophagy: cells SNARE selves.
Stroupe C. Stroupe C. Curr Biol. 2011 Sep 27;21(18):R697-9. doi: 10.1016/j.cub.2011.08.017. Curr Biol. 2011. PMID: 21959158 - Plasma membrane contributes to the formation of pre-autophagosomal structures.
Ravikumar B, Moreau K, Jahreiss L, Puri C, Rubinsztein DC. Ravikumar B, et al. Nat Cell Biol. 2010 Aug;12(8):747-57. doi: 10.1038/ncb2078. Epub 2010 Jul 18. Nat Cell Biol. 2010. PMID: 20639872 Free PMC article. - Methods to analyze SNARE-dependent vesicular fusion events that regulate autophagosome biogenesis.
Moreau K, Puri C, Rubinsztein DC. Moreau K, et al. Methods. 2015 Mar;75:19-24. doi: 10.1016/j.ymeth.2014.11.005. Epub 2014 Nov 13. Methods. 2015. PMID: 25461811 Free PMC article. - Connections between SNAREs and autophagy.
Moreau K, Renna M, Rubinsztein DC. Moreau K, et al. Trends Biochem Sci. 2013 Feb;38(2):57-63. doi: 10.1016/j.tibs.2012.11.004. Epub 2013 Jan 8. Trends Biochem Sci. 2013. PMID: 23306003 Review. - Mechanisms of autophagosome biogenesis.
Rubinsztein DC, Shpilka T, Elazar Z. Rubinsztein DC, et al. Curr Biol. 2012 Jan 10;22(1):R29-34. doi: 10.1016/j.cub.2011.11.034. Curr Biol. 2012. PMID: 22240478 Review.
Cited by
- Autophagosomal Syntaxin17-dependent lysosomal degradation maintains neuronal function in Drosophila.
Takáts S, Nagy P, Varga Á, Pircs K, Kárpáti M, Varga K, Kovács AL, Hegedűs K, Juhász G. Takáts S, et al. J Cell Biol. 2013 May 13;201(4):531-9. doi: 10.1083/jcb.201211160. J Cell Biol. 2013. PMID: 23671310 Free PMC article. - ATG14 promotes membrane tethering and fusion of autophagosomes to endolysosomes.
Diao J, Liu R, Rong Y, Zhao M, Zhang J, Lai Y, Zhou Q, Wilz LM, Li J, Vivona S, Pfuetzner RA, Brunger AT, Zhong Q. Diao J, et al. Nature. 2015 Apr 23;520(7548):563-6. doi: 10.1038/nature14147. Epub 2015 Feb 9. Nature. 2015. PMID: 25686604 Free PMC article. - Mechanism and functions of membrane binding by the Atg5-Atg12/Atg16 complex during autophagosome formation.
Romanov J, Walczak M, Ibiricu I, Schüchner S, Ogris E, Kraft C, Martens S. Romanov J, et al. EMBO J. 2012 Nov 14;31(22):4304-17. doi: 10.1038/emboj.2012.278. Epub 2012 Oct 12. EMBO J. 2012. PMID: 23064152 Free PMC article. - BRG1 attenuates colonic inflammation and tumorigenesis through autophagy-dependent oxidative stress sequestration.
Liu M, Sun T, Li N, Peng J, Fu D, Li W, Li L, Gao WQ. Liu M, et al. Nat Commun. 2019 Oct 10;10(1):4614. doi: 10.1038/s41467-019-12573-z. Nat Commun. 2019. PMID: 31601814 Free PMC article. - Emerging new roles of the lysosome and neuronal ceroid lipofuscinoses.
Mukherjee AB, Appu AP, Sadhukhan T, Casey S, Mondal A, Zhang Z, Bagh MB. Mukherjee AB, et al. Mol Neurodegener. 2019 Jan 16;14(1):4. doi: 10.1186/s13024-018-0300-6. Mol Neurodegener. 2019. PMID: 30651094 Free PMC article. Review.
References
- Axe E.L., Walker S.A., Manifava M., Chandra P., Roderick H.L., Habermann A., Griffiths G., Ktistakis N.T. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol. 2008;182:685–701. - PMC - PubMed
- Barysch S.V., Jahn R., Rizzoli S.O. A fluorescence-based in vitro assay for investigating early endosome dynamics. Nat. Protoc. 2010;5:1127–1137. - PubMed
- Bethani I., Werner A., Kadian C., Geumann U., Jahn R., Rizzoli S.O. Endosomal fusion upon SNARE knockdown is maintained by residual SNARE activity and enhanced docking. Traffic. 2009;10:1543–1559. - PubMed
- Bonifacino J.S., Glick B.S. The mechanisms of vesicle budding and fusion. Cell. 2004;116:153–166. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources