SNOSite: exploiting maximal dependence decomposition to identify cysteine S-nitrosylation with substrate site specificity - PubMed (original) (raw)
SNOSite: exploiting maximal dependence decomposition to identify cysteine S-nitrosylation with substrate site specificity
Tzong-Yi Lee et al. PLoS One. 2011.
Abstract
S-nitrosylation, the covalent attachment of a nitric oxide to (NO) the sulfur atom of cysteine, is a selective and reversible protein post-translational modification (PTM) that regulates protein activity, localization, and stability. Despite its implication in the regulation of protein functions and cell signaling, the substrate specificity of cysteine S-nitrosylation remains unknown. Based on a total of 586 experimentally identified S-nitrosylation sites from SNAP/L-cysteine-stimulated mouse endothelial cells, this work presents an informatics investigation on S-nitrosylation sites including structural factors such as the flanking amino acids composition, the accessible surface area (ASA) and physicochemical properties, i.e. positive charge and side chain interaction parameter. Due to the difficulty to obtain the conserved motifs by conventional motif analysis, maximal dependence decomposition (MDD) has been applied to obtain statistically significant conserved motifs. Support vector machine (SVM) is applied to generate predictive model for each MDD-clustered motif. According to five-fold cross-validation, the MDD-clustered SVMs could achieve an accuracy of 0.902, and provides a promising performance in an independent test set. The effectiveness of the model was demonstrated on the correct identification of previously reported S-nitrosylation sites of Bos taurus dimethylarginine dimethylaminohydrolase 1 (DDAH1) and human hemoglobin subunit beta (HBB). Finally, the MDD-clustered model was adopted to construct an effective web-based tool, named SNOSite (http://csb.cse.yzu.edu.tw/SNOSite/), for identifying S-nitrosylation sites on the uncharacterized protein sequences.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
Figure 1. The flowchart of MDD clustering.
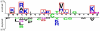
Figure 2. Frequency plot of sequence logo of S-nitrosylation sites with 21-mer window length.
Figure 3. The compositional biases of amino acids around S-nitrosylation sites compared to the non-S-nitrosylation sites.
The amino acids that are significantly enriched or depleted (_P_-value<0.05) around S-nitrosylation sites are presented.
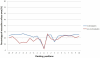
Figure 4. Comparison of average percentage of ASA in the 21-mer window (−10∼+10) between S-nitrosylation and non-S-nitrosylation sites.
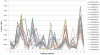
Figure 5. The top twenty physicochemical properties of S-nitrosylation sites ranked by the average value of F-score measurement in 21-mer window.
KRIW710101, side chain interaction parameter ; OOBM850105, optimized side chain interaction parameter ; FINA910104, helix termination parameter at position j+1 ; FAUJ880111, positive charge ; GUYH850104, apparent partition energies calculated from Janin index ; KIDA850101, hydrophobicity-related index ; GUYH850101, partition energy ; JANJ780101, average accessible surface area ; ROSM880102, side chain hydropathy ; CHOC760102, residue accessible surface area in folded protein ; FASG890101, hydrophobicity index ; RACS820103, average relative fractional occurrence in AL(i) ; KARP850103, flexibility parameter for two rigid neighbors ; OOBM770101, average non-bonded energy per atom ; LEVM760101, hydrophobic parameter ; MEIH800102, average reduced distance for side chain ; GUYH850105, apparent partition energies calculated from Chothia index ; KRIW790102, fraction of site occupied by water ; PUNT030101, knowledge-based membrane-propensity scale from 1D_Helix in MPtopo databases ; WEBA780101, RF value in high salt chromatography .
Figure 6. A case study of Bos taurus dimethylarginine dimethylaminohydrolase 1 (DDAH1) which contains two S-nitrosylation sites at positions 222 and 274.
Similar articles
- GSHSite: exploiting an iteratively statistical method to identify s-glutathionylation sites with substrate specificity.
Chen YJ, Lu CT, Huang KY, Wu HY, Chen YJ, Lee TY. Chen YJ, et al. PLoS One. 2015 Apr 7;10(4):e0118752. doi: 10.1371/journal.pone.0118752. eCollection 2015. PLoS One. 2015. PMID: 25849935 Free PMC article. - MDD-SOH: exploiting maximal dependence decomposition to identify S-sulfenylation sites with substrate motifs.
Bui VM, Lu CT, Ho TT, Lee TY. Bui VM, et al. Bioinformatics. 2016 Jan 15;32(2):165-72. doi: 10.1093/bioinformatics/btv558. Epub 2015 Sep 26. Bioinformatics. 2016. PMID: 26411868 - MDD-Palm: Identification of protein S-palmitoylation sites with substrate motifs based on maximal dependence decomposition.
Weng SL, Kao HJ, Huang CH, Lee TY. Weng SL, et al. PLoS One. 2017 Jun 29;12(6):e0179529. doi: 10.1371/journal.pone.0179529. eCollection 2017. PLoS One. 2017. PMID: 28662047 Free PMC article. - Strategies and tools to explore protein S-nitrosylation.
Raju K, Doulias PT, Tenopoulou M, Greene JL, Ischiropoulos H. Raju K, et al. Biochim Biophys Acta. 2012 Jun;1820(6):684-8. doi: 10.1016/j.bbagen.2011.05.009. Epub 2011 May 30. Biochim Biophys Acta. 2012. PMID: 21651963 Free PMC article. Review. - Recent Advances in Predicting Protein S-Nitrosylation Sites.
Zhao Q, Ma J, Xie F, Wang Y, Zhang Y, Li H, Sun Y, Wang L, Guo M, Han K. Zhao Q, et al. Biomed Res Int. 2021 Feb 9;2021:5542224. doi: 10.1155/2021/5542224. eCollection 2021. Biomed Res Int. 2021. PMID: 33628788 Free PMC article. Review.
Cited by
- Characterization and identification of protein O-GlcNAcylation sites with substrate specificity.
Wu HY, Lu CT, Kao HJ, Chen YJ, Chen YJ, Lee TY. Wu HY, et al. BMC Bioinformatics. 2014;15 Suppl 16(Suppl 16):S1. doi: 10.1186/1471-2105-15-S16-S1. Epub 2014 Dec 8. BMC Bioinformatics. 2014. PMID: 25521204 Free PMC article. - Investigation and identification of protein carbonylation sites based on position-specific amino acid composition and physicochemical features.
Weng SL, Huang KY, Kaunang FJ, Huang CH, Kao HJ, Chang TH, Wang HY, Lu JJ, Lee TY. Weng SL, et al. BMC Bioinformatics. 2017 Mar 14;18(Suppl 3):66. doi: 10.1186/s12859-017-1472-8. BMC Bioinformatics. 2017. PMID: 28361707 Free PMC article. - Protein S-nitrosylation: specificity and identification strategies in plants.
Lamotte O, Bertoldo JB, Besson-Bard A, Rosnoblet C, Aimé S, Hichami S, Terenzi H, Wendehenne D. Lamotte O, et al. Front Chem. 2015 Jan 7;2:114. doi: 10.3389/fchem.2014.00114. eCollection 2014. Front Chem. 2015. PMID: 25750911 Free PMC article. Review. - Protein expression profiling of nuclear membrane protein reveals potential biomarker of human hepatocellular carcinoma.
Khan R, Zahid S, Wan YJ, Forster J, Karim AB, Nawabi AM, Azhar A, Rahman MA, Ahmed N. Khan R, et al. Clin Proteomics. 2013 Jun 1;10(1):6. doi: 10.1186/1559-0275-10-6. Clin Proteomics. 2013. PMID: 23724895 Free PMC article. - ViralPhos: incorporating a recursively statistical method to predict phosphorylation sites on virus proteins.
Huang KY, Lu CT, Bretaña N, Lee TY, Chang TH. Huang KY, et al. BMC Bioinformatics. 2013;14 Suppl 16(Suppl 16):S10. doi: 10.1186/1471-2105-14-S16-S10. Epub 2013 Oct 22. BMC Bioinformatics. 2013. PMID: 24564381 Free PMC article.
References
- Bogdan C. Nitric oxide and the immune response. Nat Immun. 2001;2:907–916. - PubMed
- Karpuzoglu E, Ahmed SA. Estrogen regulation of nitric oxide and inducible nitric oxide synthase (iNOS) in immune cells: Implications for immunity, autoimmune diseases, and apoptosis. Nitric Oxide. 2006;15:177–186. - PubMed
- Stamler JS, Lamas S, Fang FC. Nitrosylation. the prototypic redox-based signaling mechanism. Cell. 2001;106:675–683. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous