CAG repeats mimic CUG repeats in the misregulation of alternative splicing - PubMed (original) (raw)
. 2011 Nov 1;39(20):8938-51.
doi: 10.1093/nar/gkr608. Epub 2011 Jul 27.
Affiliations
- PMID: 21795378
- PMCID: PMC3203611
- DOI: 10.1093/nar/gkr608
CAG repeats mimic CUG repeats in the misregulation of alternative splicing
Agnieszka Mykowska et al. Nucleic Acids Res. 2011.
Abstract
Mutant transcripts containing expanded CUG repeats in the untranslated region are a pathogenic factor in myotonic dystrophy type 1 (DM1). The mutant RNA sequesters the muscleblind-like 1 (MBNL1) splicing factor and causes misregulation of the alternative splicing of multiple genes that are linked to clinical symptoms of the disease. In this study, we show that either long untranslated CAG repeat RNA or short synthetic CAG repeats induce splicing aberrations typical of DM1. Alternative splicing defects are also caused by translated CAG repeats in normal cells transfected with a mutant ATXN3 gene construct and in cells derived from spinocerebellar ataxia type 3 and Huntington's disease patients. Splicing misregulation is unlikely to be caused by traces of antisense transcripts with CUG repeats, and the possible trigger of this misregulation may be sequestration of the MBNL1 protein with nuclear RNA inclusions containing expanded CAG repeat transcripts. We propose that alternative splicing misregulation by mutant CAG repeats may contribute to the pathological features of polyglutamine disorders.
Figures
Figure 1.
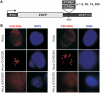
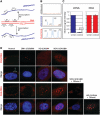
Expression of untranslated CUG and CAG repeats and cellular localization of expanded repeat transcripts. (A) Diagram of the pEGFP-N3-derived constructs used to generate stable HeLa and SK-N-MC cell lines expressing 5, 30, 74 and 200 of the CUG or CAG repeats in the 3′-UTR of mRNA encoding EGFP. (B) Representative RNA-FISH images of HeLa cells cultured in the presence of plasmid encoding (CUG)200 and (CAG)200 repeat RNAs. Nuclear retention of the mutant transcripts is visible. Nuclei were stained with DAPI (blue); RNA foci are red; magnification ×60.
Figure 2.
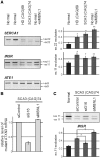
CUG and CAG repeats trigger similar pattern of the aberrant alternative splicing in HeLa cells. Total RNA isolated from cells of clonal HeLa lines expressing untranslated CUG or CAG repeat RNA of designated lengths was used to assess the levels of splicing variant transcripts from the INSR, SERCA1, CLCN1, LDB3, CAPZB, and ATE1 genes by RT–PCR. DM1-specific splicing aberrations were observed for the longest (74 and 200) both the CUG or CAG repeats and only for genes that are known to be regulated by MBNL1. The CAPZB, that is known to be regulated by CUGBP1 and ATE1 control gene that is not disrupted in DM1 patients, did not show significant changes in the proportion of transcript variants. The experiments were carried out in triplicate and quantitative results are shown as bar diagrams. The fraction of exon inclusion or exclusion (±SD) was calculated by dividing the signal of RT–PCR band corresponding to the inclusion/exclusion splice product by the total RT–PCR representing all splice products. *P = (0.05; 0.001), **P < 0.001 compared with the control.
Figure 3.
Overexpression of MBNL1 partially reverses splicing abnormalities caused by long CAG repeats in HeLa cells. Control—untransfected cells; +MBNL41 lanes—cell lines bearing long CAG tracts transfected with pEGFP-C1MBNL1 plasmid. The level of transcript expressed from pEGFP-C1MBNL1 was determined by RT–PCR analysis. The experiments were carried out in duplicate and quantitative results are shown as bar diagrams. MBNL1 overexpression was estimated in comparison to GAPDH expression level. The fraction of exon inclusion or exclusion (±SD) was calculated by dividing the signal of RT–PCR band corresponding to the inclusion/exclusion splice product by the total RT–PCR signal representing all splice products. *P = (0.05; 0.001), compared with the cells that were not transfected with pEGFP-C1MBNL1 plasmid.
Figure 4.
Short exogenous CUG and CAG repeat RNAs trigger DM1-specific splicing abnormalities in HeLa cells. RT–PCR products of the alternative splicing of MBNL1, SERCA1 and INSR genes after 48-h treatment of HeLa cells with designated synthetic oligoribonucleotides delivered in 40 nM concentration. Bar graphs show the percentage of the appropriate mRNA isoforms relative to the total amount of splice products. Splicing aberrations were observed for the CUG and CAG ORNs; (UAG)17 sequence also triggers some splicing misbalance. No changes were detected for control repeats (UAA)17, (UGG)17, siLuc, or pre-miR-136.
Figure 5.
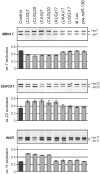
Splicing changes in cells expressing mutated Ataxin3 fragment with 69 glutamine residues. (A) Diagram of the pEGFP-mutATXN3 construct used to generate SK-N-MC cell lines expressing 69 CAG repeats in protein coding region. The stop codon of the EGFP protein was removed and a cDNA sequence of mutant human ATXN3 containing 69 CAG repeats was inserted in frame at the 3′-terminus to induce expression of EGFP-mutant Ataxin 3 fusion protein containing 69 Q-residues. (B) RT–PCR products of the alternative splicing of SERCA1, CLCN1 and control ATE1 gene. Quantification of the results is presented in the graphs as the percentage of splice products that include or exclude the indicated exons. The experiments repeated three times were carried out in three transgenic SK-N-MC lines (mutATXN3 1–3). *P = (0.05; 0.001), **P < 0.001 compared with the untreated cells (control). SK-N-MC cells with MBNL1 siRNA knocked down were used as a positive control (siMBNL1).
Figure 6.
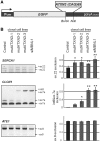
SERCA1 and INSR undergo alternative splicing changes in human HD and SCA3 fibroblasts. (A) RT–PCR analysis of endogenous SERCA1, INSR and ATE1 mRNA isoforms in fibroblasts derived from HD (69 CAG repeats in HTT) and SCA3 patients (74 CAG repeats in ATXN3) and healthy individuals (Normal). For comparison, normal cells after siRNA-mediated down-regulation of MBNL1 expression were used (+siMBNL1). A modest, but clearly visible disturbance of the balance of the SERCA1 and INSR splicing isoforms was detected in HD and SCA3 cell lines. MBNL1 knock-down resulted in analogical but stronger changes between splicing isoforms. In contrast, ATE1 mRNA expression remained unchanged. The percentages of exon 22 and exon 11 exclusion relative to the total transcripts indicate mean values ± SD of three independent experiments *P = (0.05; 0.001), **P < 0.001 compared with results obtained for normal fibroblasts). (B) The level of mutant ATXN3 transcript (74 CAG repeats) in SCA3 fibroblasts treated with three siRNAs: control, _ATXN3_-specific (siG16) and _MBNL1_-specific (siMBNL1). The relative expression level was determined by RT–PCR analysis of CAG repeat region of ATXN3 compared to GAPDH amplification product. (C) Treatment of SCA3 cells with siG16 resulted in partial reversion of INSR splicing misregulation, while MBNL1 silencing led to significant increasing of mRNA isoform without exon 11. Graph shows average values (±SD) for three independent transfection experiments [*P = (0.05; 0.001)].
Figure 7.
Identification of sense and antisense repeat transcripts in cells expressing long CUG and CAG repeats. (A) Schematic representation of strand-specific MLPA assay design with probes located in the EGFP sequence close to the position of the changeable repeat tract. Signal from the probes hybridizing to plasmid DNA represents equal dosage from sense and antisense strands. The reverse transcription (RT) was performed with DNase treated total RNA. The signal from probe hybridizing to cDNA representing sense and antisense (a-sense) transcripts is proportional to the cellular level of these transcripts. (B) Representative MLPA electrophoregrams show results obtained for: cDNA from untransfected cells where no signals from sense and antisense probes are detected (top panel), cDNA from cells transfected with pEGFP-derived plasmid encoding (CAG)5 where very low, barely detectable signals from antisense-specific probes (A1 and A2) are visible (middle panel) and pEGFP-N3 plasmid DNA, where comparable signals from both the sense- and the antisense-specific probes are shown (lower panel). The signal from sense (S1 and S2) and antisense (A1 and A2) probes are indicated. (C) Bar plots show fraction of signal from the antisense-specific probes normalized to average signal from the sense-specific probe obtained from five cDNA samples of SK-N-MC cells transfected with the pEGFP-N3 encoding: (CAG)5, (CAG)74, (CUG)5 and (CUG)74 and mutATXN3 (left-hand side) and from two samples of pEGFP-N3 plasmid DNA (right-hand side). (D) Representative images of RNA-FISH in cultured human fibroblasts using Cy3-labeled DNA/LNA probes (CAG)6-CA (upper rows) and (CTG)6-CA (lower rows). HD cells expressing 44 CAG repeats and SCA3 cells expressing 69 CAG repeats from, respectively, the HTT and the ATNX3 genes show nuclear retention of sense transcripts harboring the CAG repeat expansions. Such CAG RNA inclusions were not detected in the nuclei of normal and DM1 fibroblasts when FISH was carried out with the same CTG probe. The presence of CAG repeat RNA in the nuclear inclusions was confirmed by RNase A (no FISH signal) and by DNaseI treatment (presence of FISH signal). The antisense transcripts through the repeat region in mutant HTT and ATXN3 genes were not detected with CAG probe in the nuclear RNA inclusions of HD and SCA3 cells. In this assay, DM1 cells were included as a positive control to detect ribonuclear CUG repeat inclusions. CAG and CUG nuclear RNA foci (red); DAPI nuclear stain (blue).
Figure 8.
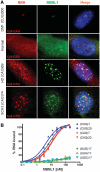
MBNL1 protein binds CAG and CUG repeats in vitro and co-localizes with repeat region of mutant HTT and ATXN3 transcripts in nuclear inclusions. (A) Combined FISH/IF assay using Cy3-labeled CAG probe in DM1 cells and CTG probe in normal, HD and SCA3 cells as well as rabbit polyclonal MBNL1 primary antibody (A2764) detected with secondary Cy2-labeled anti-rabbit antibody. Merged images show the colocalization of the MBNL1 protein with nuclear CAG repeat RNA inclusions in HD and SCA3 cells. Such colocalization is also demonstrated in DM1 cells with CUG RNA foci. In normal fibroblasts, MBNL1 is detected throughout the cell. DAPI staining (blue), MBNL1 (green), CAG and CUG nuclear RNA (red). (B) Results of filter binding assay to determine binding affinity of MBNL1 protein to 5′-end labeled transcripts of (CUG)7, (CUG)20, (CAG)7, (CAG)20, (AUG)17, (UUA)17 and (UGG)17. Affinity of MBNL1 to CUG and CAG RNA is similar with only slight preference for the CUG repeats.
Similar articles
- RNA FISH for detecting expanded repeats in human diseases.
Urbanek MO, Krzyzosiak WJ. Urbanek MO, et al. Methods. 2016 Apr 1;98:115-123. doi: 10.1016/j.ymeth.2015.11.017. Epub 2015 Nov 23. Methods. 2016. PMID: 26615955 - Colocalization of muscleblind with RNA foci is separable from mis-regulation of alternative splicing in myotonic dystrophy.
Ho TH, Savkur RS, Poulos MG, Mancini MA, Swanson MS, Cooper TA. Ho TH, et al. J Cell Sci. 2005 Jul 1;118(Pt 13):2923-33. doi: 10.1242/jcs.02404. Epub 2005 Jun 16. J Cell Sci. 2005. PMID: 15961406 - Short antisense-locked nucleic acids (all-LNAs) correct alternative splicing abnormalities in myotonic dystrophy.
Wojtkowiak-Szlachcic A, Taylor K, Stepniak-Konieczna E, Sznajder LJ, Mykowska A, Sroka J, Thornton CA, Sobczak K. Wojtkowiak-Szlachcic A, et al. Nucleic Acids Res. 2015 Mar 31;43(6):3318-31. doi: 10.1093/nar/gkv163. Epub 2015 Mar 9. Nucleic Acids Res. 2015. PMID: 25753670 Free PMC article. - Gain of RNA function in pathological cases: Focus on myotonic dystrophy.
Klein AF, Gasnier E, Furling D. Klein AF, et al. Biochimie. 2011 Nov;93(11):2006-12. doi: 10.1016/j.biochi.2011.06.028. Epub 2011 Jul 13. Biochimie. 2011. PMID: 21763392 Review. - An Overview of Alternative Splicing Defects Implicated in Myotonic Dystrophy Type I.
López-Martínez A, Soblechero-Martín P, de-la-Puente-Ovejero L, Nogales-Gadea G, Arechavala-Gomeza V. López-Martínez A, et al. Genes (Basel). 2020 Sep 22;11(9):1109. doi: 10.3390/genes11091109. Genes (Basel). 2020. PMID: 32971903 Free PMC article. Review.
Cited by
- Twisting right to left: A…A mismatch in a CAG trinucleotide repeat overexpansion provokes left-handed Z-DNA conformation.
Khan N, Kolimi N, Rathinavelan T. Khan N, et al. PLoS Comput Biol. 2015 Apr 13;11(4):e1004162. doi: 10.1371/journal.pcbi.1004162. eCollection 2015 Apr. PLoS Comput Biol. 2015. PMID: 25876062 Free PMC article. - Examining the interactions of the splicing factor MBNL1 with target RNA sequences via a label-free, multiplex method.
Yadav AR, Mace CR, Miller BL. Yadav AR, et al. Anal Chem. 2014 Jan 21;86(2):1067-75. doi: 10.1021/ac402603j. Epub 2014 Jan 9. Anal Chem. 2014. PMID: 24377303 Free PMC article. - Design of Bivalent Nucleic Acid Ligands for Recognition of RNA-Repeated Expansion Associated with Huntington's Disease.
Thadke SA, Perera JDR, Hridya VM, Bhatt K, Shaikh AY, Hsieh WC, Chen M, Gayathri C, Gil RR, Rule GS, Mukherjee A, Thornton CA, Ly DH. Thadke SA, et al. Biochemistry. 2018 Apr 10;57(14):2094-2108. doi: 10.1021/acs.biochem.8b00062. Epub 2018 Mar 27. Biochemistry. 2018. PMID: 29562132 Free PMC article. - Role and Perspective of Molecular Simulation-Based Investigation of RNA-Ligand Interaction: From Small Molecules and Peptides to Photoswitchable RNA Binding.
Berdnikova DV, Carloni P, Krauß S, Rossetti G. Berdnikova DV, et al. Molecules. 2021 Jun 3;26(11):3384. doi: 10.3390/molecules26113384. Molecules. 2021. PMID: 34205049 Free PMC article. Review. - Alterations in mRNA 3' UTR Isoform Abundance Accompany Gene Expression Changes in Human Huntington's Disease Brains.
Romo L, Ashar-Patel A, Pfister E, Aronin N. Romo L, et al. Cell Rep. 2017 Sep 26;20(13):3057-3070. doi: 10.1016/j.celrep.2017.09.009. Cell Rep. 2017. PMID: 28954224 Free PMC article.
References
- Bauer PO, Nukina N. The pathogenic mechanisms of polyglutamine diseases and current therapeutic strategies. J. Neurochem. 2009;110:1737–1765. - PubMed
- de Leon MB, Cisneros B. Myotonic dystrophy 1 in the nervous system: from the clinic to molecular mechanisms. J. Neurosci. Res. 2008;86:18–26. - PubMed
- Underwood BR, Rubinsztein DC. Spinocerebellar ataxias caused by polyglutamine expansions: a review of therapeutic strategies. Cerebellum. 2008;7:215–221. - PubMed
- Galvao R, Mendes-Soares L, Camara J, Jaco I, Carmo-Fonseca M. Triplet repeats, RNA secondary structure and toxic gain-of-function models for pathogenesis. Brain Res. Bull. 2001;56:191–201. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources