Spatial distribution of microbial communities in the cystic fibrosis lung - PubMed (original) (raw)
Spatial distribution of microbial communities in the cystic fibrosis lung
Dana Willner et al. ISME J. 2012 Feb.
Abstract
Cystic fibrosis (CF) is a common fatal genetic disorder with mortality most often resulting from microbial infections of the lungs. Culture-independent studies of CF-associated microbial communities have indicated that microbial diversity in the CF airways is much higher than suggested by culturing alone. However, these studies have relied on indirect methods to sample the CF lung such as expectorated sputum and bronchoalveolar lavage (BAL). Here, we characterize the diversity of microbial communities in tissue sections from anatomically distinct regions of the CF lung using barcoded 16S amplicon pyrosequencing. Microbial communities differed significantly between different areas of the lungs, and few taxa were common to microbial communities in all anatomical regions surveyed. Our results indicate that CF lung infections are not only polymicrobial, but also spatially heterogeneous suggesting that treatment regimes tailored to dominant populations in sputum or BAL samples may be ineffective against infections in some areas of the lung.
Figures
Figure 1
Relative abundance of microbial OTUs in the ex-plant and postmortem lungs. Each row represents a different OTU, and the abundance as a percentage of the total population is indicated by color. Phyla are indicated on the left, and a subset of OTU classifications is indicated on the right. A complete list of OTU classification appears in Supplementary Table 2. Lung regions are abbreviated as RUL for right upper lobe, RML for right middle lobe, RLL for right lower lobe, LUL for left upper lobe, Ling for left lingula and LLL for left lower lobe. LLLA indicates the left lower lobe anterior and LLLP indicates the left lower lobe posterior. Trach indicates the tracheal sample.
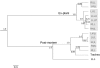
Figure 2
Spatial clustering of microbial communities in the CF lungs. Communities were compared using the weighted Unifrac distance metric, and clustered using average linkage. Node labels indicate jackknife support values, and the bar below the tree represents a weighted Unifrac distance of 0.01. Shaded boxes indicate communities that were not significantly different from each other, but were significantly different from all other communities as determined by XIPE.
Similar articles
- Analysis of Lung Microbiota in Bronchoalveolar Lavage, Protected Brush and Sputum Samples from Subjects with Mild-To-Moderate Cystic Fibrosis Lung Disease.
Hogan DA, Willger SD, Dolben EL, Hampton TH, Stanton BA, Morrison HG, Sogin ML, Czum J, Ashare A. Hogan DA, et al. PLoS One. 2016 Mar 4;11(3):e0149998. doi: 10.1371/journal.pone.0149998. eCollection 2016. PLoS One. 2016. PMID: 26943329 Free PMC article. - Different next generation sequencing platforms produce different microbial profiles and diversity in cystic fibrosis sputum.
Hahn A, Sanyal A, Perez GF, Colberg-Poley AM, Campos J, Rose MC, Pérez-Losada M. Hahn A, et al. J Microbiol Methods. 2016 Nov;130:95-99. doi: 10.1016/j.mimet.2016.09.002. Epub 2016 Sep 5. J Microbiol Methods. 2016. PMID: 27609714 Free PMC article. - Upper versus lower airway microbiome and metagenome in children with cystic fibrosis and their correlation with lung inflammation.
Kirst ME, Baker D, Li E, Abu-Hasan M, Wang GP. Kirst ME, et al. PLoS One. 2019 Sep 19;14(9):e0222323. doi: 10.1371/journal.pone.0222323. eCollection 2019. PLoS One. 2019. PMID: 31536536 Free PMC article. - Directly sampling the lung of a young child with cystic fibrosis reveals diverse microbiota.
Brown PS, Pope CE, Marsh RL, Qin X, McNamara S, Gibson R, Burns JL, Deutsch G, Hoffman LR. Brown PS, et al. Ann Am Thorac Soc. 2014 Sep;11(7):1049-55. doi: 10.1513/AnnalsATS.201311-383OC. Ann Am Thorac Soc. 2014. PMID: 25072206 Free PMC article. Review. - Same Game, Different Players: Emerging Pathogens of the CF Lung.
Gannon AD, Darch SE. Gannon AD, et al. mBio. 2021 Jan 12;12(1):e01217-20. doi: 10.1128/mBio.01217-20. mBio. 2021. PMID: 33436426 Free PMC article. Review.
Cited by
- High individuality of respiratory bacterial communities in a large cohort of adult cystic fibrosis patients under continuous antibiotic treatment.
Kramer R, Sauer-Heilborn A, Welte T, Jauregui R, Brettar I, Guzman CA, Höfle MG. Kramer R, et al. PLoS One. 2015 Feb 11;10(2):e0117436. doi: 10.1371/journal.pone.0117436. eCollection 2015. PLoS One. 2015. PMID: 25671713 Free PMC article. - Ability of device to collect bacteria from cough aerosols generated by adults with cystic fibrosis.
Ku DN, Ku SK, Helfman B, McCarty NA, Wolff BJ, Winchell JM, Anderson LJ. Ku DN, et al. F1000Res. 2016 Aug 5;5:1920. doi: 10.12688/f1000research.9251.1. eCollection 2016. F1000Res. 2016. PMID: 27781088 Free PMC article. - Variability in Bacteriophage and Antibiotic Sensitivity in Serial Pseudomonas aeruginosa Isolates from Cystic Fibrosis Airway Cultures over 12 Months.
Martin I, Kenna DTD, Morales S, Alton EWFW, Davies JC. Martin I, et al. Microorganisms. 2021 Mar 22;9(3):660. doi: 10.3390/microorganisms9030660. Microorganisms. 2021. PMID: 33810202 Free PMC article. - mSphere of Influence: a Community To Study Communities.
Limoli D. Limoli D. mSphere. 2020 Feb 5;5(1):e00047-20. doi: 10.1128/mSphere.00047-20. mSphere. 2020. PMID: 32024707 Free PMC article. - Cystic fibrosis: NHLBI Workshop on the Primary Prevention of Chronic Lung Diseases.
Pittman JE, Cutting G, Davis SD, Ferkol T, Boucher R. Pittman JE, et al. Ann Am Thorac Soc. 2014 Apr;11 Suppl 3(Suppl 3):S161-8. doi: 10.1513/AnnalsATS.201312-444LD. Ann Am Thorac Soc. 2014. PMID: 24754825 Free PMC article. Review.
References
- Armougom F, Bittar F, Stremler N, Rolain JM, Robert C, Dubus JC, et al. Microbial diversity in the sputum of a cystic fibrosis patient studied with 16S rDNA pyrosequencing. Eur J Clin Microbiol Infect Dis. 2009;28:1151–1154. - PubMed
- Bittar EE. Pulmonary Biol health Dis. Springer; 2002.