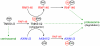Ubiquitin ligase RNF146 regulates tankyrase and Axin to promote Wnt signaling - PubMed (original) (raw)
doi: 10.1371/journal.pone.0022595. Epub 2011 Jul 25.
Hoanh Tran, Lilian Phu, Ted Lau, James Lee, Wendy N Sandoval, Peter S Liu, Sheila Bheddah, Janet Tao, Jennie R Lill, Jo-Anne Hongo, David Davis, Donald S Kirkpatrick, Paul Polakis, Mike Costa
Affiliations
- PMID: 21799911
- PMCID: PMC3143158
- DOI: 10.1371/journal.pone.0022595
Ubiquitin ligase RNF146 regulates tankyrase and Axin to promote Wnt signaling
Marinella G Callow et al. PLoS One. 2011.
Abstract
Canonical Wnt signaling is controlled intracellularly by the level of β-catenin protein, which is dependent on Axin scaffolding of a complex that phosphorylates β-catenin to target it for ubiquitylation and proteasomal degradation. This function of Axin is counteracted through relocalization of Axin protein to the Wnt receptor complex to allow for ligand-activated Wnt signaling. AXIN1 and AXIN2 protein levels are regulated by tankyrase-mediated poly(ADP-ribosyl)ation (PARsylation), which destabilizes Axin and promotes signaling. Mechanistically, how tankyrase limits Axin protein accumulation, and how tankyrase levels and activity are regulated for this function, are currently under investigation. By RNAi screening, we identified the RNF146 RING-type ubiquitin E3 ligase as a positive regulator of Wnt signaling that operates with tankyrase to maintain low steady-state levels of Axin proteins. RNF146 also destabilizes tankyrases TNKS1 and TNKS2 proteins and, in a reciprocal relationship, tankyrase activity reduces RNF146 protein levels. We show that RNF146, tankyrase, and Axin form a protein complex, and that RNF146 mediates ubiquitylation of all three proteins to target them for proteasomal degradation. RNF146 is a cytoplasmic protein that also prevents tankyrase protein aggregation at a centrosomal location. Tankyrase auto-PARsylation and PARsylation of Axin is known to lead to proteasome-mediated degradation of these proteins, and we demonstrate that, through ubiquitylation, RNF146 mediates this process to regulate Wnt signaling.
Conflict of interest statement
Competing Interests: The authors have read the journal's policy and have the following conflicts. All authors are employed by Genentech Research and Early Development. There are no patents, products in development, or marketed products to declare for this manuscript. This does not alter the authors' adherence to all the PLoS ONE policies on sharing data and materials.
Figures
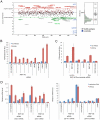
Figure 1. RNF146 positively regulates Wnt signaling.
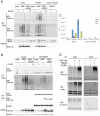
(A) Scatter plot of Z-scores for the RNAi screen of human E3 ligase siRNA pools in Wnt3a-stimulated HEK293 cells stably expressing dual luciferase reporters . Z-scores on the y axis represent TOPbrite Wnt reporter values normalized to SV40 reporter values. Positively and negatively regulating siRNA pools that deviate from the Z-score cut-off lines are shown in green and red, respectively, by gene name. Control siRNAs targeting CTNNB1 (β-catenin), LRP6 and APC, as well as control wells not induced with Wnt3a protein, are depicted in blue but were excluded from the distribution analysis. SV40 reporter values are plotted to the right, with outliers deviating from the normal distribution listed in gray on both graphs. (B) RNF146 RNAi by transient transfection of four individual and pooled siRNAs in HEK293 cells showing inhibition of the Wnt3a response (red) and lack of nonspecific effects on uninduced (blue) reporter activity. Inhibition by LRP6 and β-catenin siRNAs, and activation by AXIN1 and AXIN2 siRNAs, are shown as controls. Error bars in this and all figures represent the standard deviation of at least three replicate samples. (C) RNF146 RNAi in HEK293T cells stably expressing doxycycline (Dox)-inducible miRNA targeting RNF146, and transiently transfected with TOPbrite Wnt luciferase reporter with or without wildtype or dominant-negative (H53A) RNF146 expression constructs. (D) qRT (quantitative real-time)-PCR mRNA expression analysis of Wnt target genes in PA-1 cells transfected with RNF146, LRP6 (positive control), and non-targeting (negative control) siRNAs. Cells were either unstimulated with exogenous Wnt3a (blue) to test effects on autocrine Wnt signaling , or further induced with Wnt3a (red).
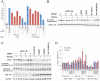
Figure 2. RNF146 acts in the Wnt pathway at the level of Axin protein destabilization.
(A) Effects of individual RNF146 (red) and control (blue) LRP6, β-catenin, or non-targeting siRNAs on Wnt reporter activity in HEK293 cells stably expressing TOPbrite and SV40 reporters, and transiently transfected with expression construct for either wildtype LRP6 or a constitutively active LRP6 with the extracellular domain deleted (deltaN). (B) Western analysis of the effects of individual RNF146 siRNAs on β-catenin stabilization induced by Wnt3a (+) in HEK293 cells. LRP5 and LRP6 siRNAs are used as negative and positive controls, respectively, since Wnt signaling in HEK293 cells depends on LRP6 and not LRP5 . β-catenin siRNA indicates that the higher molecular weight band in the panel corresponds to β-catenin protein. Clathrin heavy chain immunoblotting was used as a gel loading control. (C) Western analysis of AXIN1, RNF146, β-catenin phosphoshorylated on Ser33, Ser37, and Thr41, and active GSK3α/β auto-phosphorylated on Tyr279/216 after transfection of individual RNF146 siRNAs in HEK293 cells with (+) or without (−) Wnt3a stimulation. β-catenin and AXIN1 RNAi confirm the specificity of the antibodies. GAPDH was used as a loading control. (D) qRT-PCR analysis of AXIN1, AXIN2, and RNF146 mRNA after transfection of individual RNF146 siRNAs in HEK293 cells. Note that AXIN2 expression is weakly induced by Wnt3a (red), and this response is regulated by the RNAi treatments.
Figure 3. RNF146 and tankyrase function coordinately in Wnt signaling and regulate each other's protein stability.
(A) Effects on Wnt signaling in the HEK293 reporter stable cell line with and without Wnt3a stimulation for siRNAs targeting RNF146, the combination of tankyrases TNKS1 and TNKS2, or the combination of all three genes. On the right half of the graph, these siRNAs are also combined with AXIN1 siRNA. AXIN2, β-catenin, and non-targeting siRNAs serve as controls. (B) Western analysis of whole cell lysates for HEK293 cells treated as in (A). (C) Time course of tankyrase (TNKS1 and TNKS2) and Axin stabilization in HEK293T cells stably expressing RNF146 miRNA that was induced for the indicated times with doxycycline (DOX), with or without Wnt3a added for the final 12 h of DOX induction. β-catenin phosphorylation and RNF146 protein knockdown levels are also shown. GSK3α/βimmunoblotting was used as a loading control. (D) Western analysis of HEK293 cells treated with tankyrase small-molecule inhibitor IWR-2 at the indicated concentrations for 16 h without (−) or with (+) Wnt3a induction. Whole cell lysates were monitored for AXIN1, RNF146, and β-catenin protein levels, and GAPDH serves as a loading control. Transgenic overexpression of RNF46 partially blocks AXIN1 protein stabilization by IWR-2 compound (right-most two lanes). (E) Wnt reporter activity in the HEK293 stable cell line transfected with expression constructs for the indicated RNF146 alleles, co-expressed with either control vector (blue, without Wnt3a stimulation, or red, with Wnt3a) or RNF146 (green, without Wnt3a, or purple, with Wnt3a). A cell viability assay was used to normalize Wnt luciferase reporter activity . (F) Corresponding Western analysis of Axin, RNF146, and tankyrase proteins for cells treated as in (E). (G) Schematic representation of the RNF146 proteins produced by the expression constructs, with structural domains and mutation sites indicated. (H) Western analysis of endogenous tankyrase and Axin protein levels in HEK293T cells stably expressing doxycycline-induced RNF146 (RNF) or control (lacZ) miRNA after transfection with the indicated RNF146 or control vector expression plasmids, without (−) or with (+) Wnt3a induction. Anti-HA immunoblotting shows the expression level and RNAi-mediated knockdown of HA-RNF146 protein, and GAPDH levels serve as a loading control.
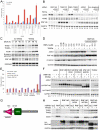
Figure 4. RNF146 RNAi does not inhibit Wnt signaling in β-catenin or APC mutant colorectal cell lines.
(A) Expression levels of endogenous β-catenin-activated genes AXIN2 (blue) and SP5 (red) in SW48 cells after transfection of individual or pooled RNF146 siRNAs. β-catenin and non-targeting siRNAs serve as positive and negative controls, respectively, and the level of knockdown of RNF146 mRNA expression is indicated in green. (B) Co-transfection of RNF146 (red), tankyrase (both TNKS1 and TNKS2; green), or control non-targeting siRNA with β-catenin siRNA at the indicated concentration in HCT-15 cells stably expressing TOPbrite Wnt reporter. Reporter activity is normalized to cell number and non-targeting siRNA treatment alone. (C–D) Western analysis of tankyrase, Axin, and RNF146 protein levels in HCT-15 (C) or SW480 (D) cells after siRNA treatment targeting the indicated genes.
Figure 5. RNF146 displays ubiquitin E3 ligase activity in vitro and binds to tankyrase, PARP1, and Axin proteins.
(A) Western analysis of auto-ubiquitylation reactions with variable amounts of GST-RNF146 protein (see Materials and Methods) immunoblotted for flag-ubiquitin, polyubiquitin, or GST. GST protein serves as a negative control. (B) Western analysis as in (A) for GST-RNF146 ubiquitylation reactions incubated in the absence (−) or presence (+) of poly(ADP-ribose) [PAR] and immunoprecipitated with antibodies specific for K11-, K48-, or K63-linked polyubiquitin. Immunoblotting with RNF146 antibody serves as a loading control. Note that the K48 linkage-specific antibody more efficiently immunoprecipitates ubiquitylated RNF146, although the polyubiquitin chains are shorter in length and therefore less readily detected by anti-flag or -ubiquitin immunoblotting. (C) Coomassie-stained gel of anti-HA immunoprecipitates from cells transfected with the indicated expression constructs for RNF146 or control E3 ligase HECTD1. Numbered protein bands and lettered high-molecular-weight bands were excised for mass spectrometric analysis. (D) Table of proteins identified from the mass spectrometric analysis showing total numbers of peptides identified for each protein, combined for all numbered protein bands in (C). Shown are 23 proteins with the highest number of total peptides identified by interaction with RNF146ΔRING protein, and with fewer than three peptides in the spectrometric analysis of HECTD1 protein interactors. The second set of 5 proteins in the table show the greatest numbers of peptides identified for the analysis of interactors with wildtype RNF146 protein, but not HECTD1 protein. The code for the coloring is explained in the legend. (E) Western analysis of anti-V5 immunoprecipitation from HEK293 cells co-transfected as indicated for expression of V5-tagged wildtype or H53A mutant RNF146 (RNF), HA-tagged TNKS2, or control vector in the presence (+) and absence (−) of proteasome inhibitor ALLN. Co-immunoprecipitation of endogenous TNKS1 and overexpressed TNKS2 was assessed with anti-TNKS1/2 antibodies from the indicated two sources. RNF146 immunoblotting, anti-HA immunoprecipitation, and whole cell lysates are shown as controls. (F) Western analysis of immunoprecipitation of flag-tagged AXIN1 expressed in HEK293 cells co-transfected with the indicated expression constructs for HA-tagged wildtype or deletion mutant alleles of RNF146, TNKS1, or PARP1. Short exposure to film of the anti-HA immunoblot detects co-immunoprecipitated TNKS1 proteins, whereas longer exposure reveals RNF146 and PARP1 proteins. Input whole cell lysates probed for HA and flag detection are shown as controls for expression of the indicated proteins.
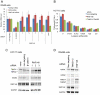
Figure 6. RNF146 ubiquitylates tankyrase and Axin in cells.
(A) Western analysis of anti-ubiquitin (FK2 antibody) immunoprecipitation from HEK293 cells transfected with expression constructs for wildtype or H53A mutant RNF146 (RNF) or control E3 ligase AMFR, in combination with either TNKS2 or control vector DNA, without (−) or with (+) proteasome inhibitor ALLN treatment. Immunoblots are shown for ubiquitylated tankyrase and RNF146 smears. Input lysate immunoblots show expression levels of tankyrase and RNF146. (B) Western analysis analogous to (A) except that HA-tagged PARP1 is overexpressed rather that TNKS2. RNF146 binding to PARP1 is confirmed by anti-HA immunoprecipitation. (C) Relative quantitation of the area under the mass spectra curve (AUC) for K48-, K63-, and K11-linked polyubiquitin -GG signature peptides in excised gel bands A, B, and C depicted in Figure 5C. Results are shown for immunoprecipitation of wildtype or H53A mutant RNF146 proteins expressed in HEK293 cells. (D) Western analysis of immunoprecipitation with K48 or K63 linkage-specific polyubiquitin antibodies for the indicated overexpression of RNF146, tankyrase, or Axin in HEK293 cells. Immunoblotting for tankyrase, Axin, or control β-catenin proteins detects high-molecular-weight polyubiquitylated protein species.
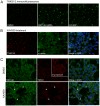
Figure 7. RNF146 RNAi and tankyrase inhibition induce tankyrase puncta in cells.
(A) Immunofluorescence imaging of tankyrase (green) in HEK293T cells stably expressing doxycycline-inducible RNF146 miRNA treated with either control DMSO, doxycycline (RNF146 miRNA), or tankyrase inhibitor XAV939. (B) HEK293 cells treated with XAV939 and immunostained for endogenous tankyrase (red) and γ-tubulin (green). The merged image shows co-localized tankyrase with γ-tubulin (yellow) counterstained for nuclei with DAPI (blue). All images are representative of at least three independent experiments. (C) HEK293 cells were treated with DMSO or XAV939 and immunostained for endogenous RNF146 (green) and tankyrase (red), with DAPI counterstaining (blue) in the merged image. Arrowheads indicate co-localization of RNF146 and tankyrase in puncta.
Figure 8. Model of RNF146 activity that leads to degradation of tankyrase, Axin, and RNF146 proteins.
See Discussion for details. Green arrows indicate protein relocalization. “Ub” indicates ubiquitylation and “PAR” indicates PARsylation of the indicated proteins.
Similar articles
- RNF146 is a poly(ADP-ribose)-directed E3 ligase that regulates axin degradation and Wnt signalling.
Zhang Y, Liu S, Mickanin C, Feng Y, Charlat O, Michaud GA, Schirle M, Shi X, Hild M, Bauer A, Myer VE, Finan PM, Porter JA, Huang SM, Cong F. Zhang Y, et al. Nat Cell Biol. 2011 May;13(5):623-9. doi: 10.1038/ncb2222. Epub 2011 Apr 10. Nat Cell Biol. 2011. PMID: 21478859 - A Context-Dependent Role for the RNF146 Ubiquitin Ligase in Wingless/Wnt Signaling in Drosophila.
Wang Z, Tacchelly-Benites O, Noble GP, Johnson MK, Gagné JP, Poirier GG, Ahmed Y. Wang Z, et al. Genetics. 2019 Mar;211(3):913-923. doi: 10.1534/genetics.118.301393. Epub 2018 Dec 28. Genetics. 2019. PMID: 30593492 Free PMC article. - Computational study on cross-talking cancer signalling mechanism of ring finger protein 146, AXIN and Tankyrase protein complex.
Loganathan L, Natarajan K, Muthusamy K. Loganathan L, et al. J Biomol Struct Dyn. 2020 Oct;38(17):5173-5185. doi: 10.1080/07391102.2019.1696707. Epub 2019 Dec 2. J Biomol Struct Dyn. 2020. PMID: 31760854 - PARsylation-mediated ubiquitylation: lessons from rare hereditary disease Cherubism.
Matsumoto Y, Rottapel R. Matsumoto Y, et al. Trends Mol Med. 2023 May;29(5):390-405. doi: 10.1016/j.molmed.2023.02.001. Epub 2023 Mar 20. Trends Mol Med. 2023. PMID: 36948987 Review. - Ring finger protein 146/Iduna is a poly(ADP-ribose) polymer binding and PARsylation dependent E3 ubiquitin ligase.
Zhou ZD, Chan CH, Xiao ZC, Tan EK. Zhou ZD, et al. Cell Adh Migr. 2011 Nov-Dec;5(6):463-71. doi: 10.4161/cam.5.6.18356. Cell Adh Migr. 2011. PMID: 22274711 Free PMC article. Review.
Cited by
- Tankyrases as modulators of pro-tumoral functions: molecular insights and therapeutic opportunities.
Zamudio-Martinez E, Herrera-Campos AB, Muñoz A, Rodríguez-Vargas JM, Oliver FJ. Zamudio-Martinez E, et al. J Exp Clin Cancer Res. 2021 Apr 28;40(1):144. doi: 10.1186/s13046-021-01950-6. J Exp Clin Cancer Res. 2021. PMID: 33910596 Free PMC article. Review. - The Colossus of ubiquitylation: decrypting a cellular code.
Williamson A, Werner A, Rape M. Williamson A, et al. Mol Cell. 2013 Feb 21;49(4):591-600. doi: 10.1016/j.molcel.2013.01.028. Mol Cell. 2013. PMID: 23438855 Free PMC article. - Gtpbp2 is a positive regulator of Wnt signaling and maintains low levels of the Wnt negative regulator Axin.
Gillis WQ, Kirmizitas A, Iwasaki Y, Ki DH, Wyrick JM, Thomsen GH. Gillis WQ, et al. Cell Commun Signal. 2016 Aug 2;14(1):15. doi: 10.1186/s12964-016-0138-x. Cell Commun Signal. 2016. PMID: 27484226 Free PMC article. - The way Wnt works: components and mechanism.
Saito-Diaz K, Chen TW, Wang X, Thorne CA, Wallace HA, Page-McCaw A, Lee E. Saito-Diaz K, et al. Growth Factors. 2013 Feb;31(1):1-31. doi: 10.3109/08977194.2012.752737. Epub 2012 Dec 21. Growth Factors. 2013. PMID: 23256519 Free PMC article. Review. - Proteome-wide Analysis Reveals Substrates of E3 Ligase RNF146 Targeted for Degradation.
Nie L, Wang C, Li N, Feng X, Lee N, Su D, Tang M, Yao F, Chen J. Nie L, et al. Mol Cell Proteomics. 2020 Dec;19(12):2015-2030. doi: 10.1074/mcp.RA120.002290. Epub 2020 Sep 21. Mol Cell Proteomics. 2020. PMID: 32958691 Free PMC article.
References
- Giles RH, van Es JH, Clevers H. Caught up in a Wnt storm: Wnt signaling in cancer. Biochim Biophys Acta. 2003;1653:1–24. - PubMed
- Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. 2004;20:781–810. - PubMed
- Salic A, Lee E, Mayer L, Kirschner MW. Control of beta-catenin stability: reconstitution of the cytoplasmic steps of the Wnt pathway in Xenopus egg extracts.. Mol Cell. 2000;5:523–32. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases