Impact of small repeat sequences on bacterial genome evolution - PubMed (original) (raw)
Impact of small repeat sequences on bacterial genome evolution
Nicholas Delihas. Genome Biol Evol. 2011.
Abstract
Intergenic regions of prokaryotic genomes carry multiple copies of terminal inverted repeat (TIR) sequences, the nonautonomous miniature inverted-repeat transposable element (MITE). In addition, there are the repetitive extragenic palindromic (REP) sequences that fold into a small stem loop rich in G-C bonding. And the clustered regularly interspaced short palindromic repeats (CRISPRs) display similar small stem loops but are an integral part of a complex genetic element. Other classes of repeats such as the REP2 element do not have TIRs but show other signatures. With the current availability of a large number of whole-genome sequences, many new repeat elements have been discovered. These sequences display diverse properties. Some show an intimate linkage to integrons, and at least one encodes a small RNA. Many repeats are found fused with chromosomal open reading frames, and some are located within protein coding sequences. Small repeat units appear to work hand in hand with the transcriptional and/or post-transcriptional apparatus of the cell. Functionally, they are multifaceted, and this can range from the control of gene expression, the facilitation of host/pathogen interactions, or stimulation of the mammalian immune system. The CRISPR complex displays dramatic functions such as an acquired immune system that defends against invading viruses and plasmids. Evolutionarily, mobile repeat elements may have influenced a cycle of active versus inactive genes in ancestral organisms, and some repeats are concentrated in regions of the chromosome where there is significant genomic plasticity. Changes in the abundance of genomic repeats during the evolution of an organism may have resulted in a benefit to the cell or posed a disadvantage, and some present day species may reflect a purification process. The diverse structure, eclectic functions, and evolutionary aspects of repeat elements are described.
Figures
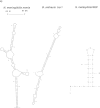
FIG. 1.—
(a) Predicted secondary structures of repeat sequences at the RNA level. The Mfold program was used for RNA folding (Markham and Zuker 2005). The Neisseria meningitidis nemis (neisseria miniature ISs) is characteristic of MITEs, and the secondary structure shown is similar to that of Mazzone et al. (2001). The top schematic describes inverted repeats (IR) and DRs flanking the DNA strand. The bcr1 structure is that of Bacillus anthracis 1R (Økstad et al. 2004) and is typical of the Bacillus bcr1 RNA secondary structures (Klevan et al. 2007). These consist of a cruciform-like structure with two independent stem loops. The Stenotrophomonas maltophilia REP sequence and secondary structure shown is characteristic of the short high G–C content REPs found in these species; they are termed SMAG (Rocco et al. 2010). These SMAG units can carry an unpaired tetranucleotide sequence at one end. (b) Left, predicted RNA secondary structures of the REP2 sequence from N. meningitidis showing internal stem loops 1 and 2. The nt sequence is from Morelle et al. (2003). Upper schematic denotes the REP2 DNA strand with promoter, ribosome binding site (RBS), and ATG initiation codon. (b) Right, predicted secondary structural model of the Borrelia burgdoferi IR-A sequence from circular plasmid cp8.3/Ip21 [nt sequence from Dunn et al. (1994)]. Stem loops 1 and 2 may be analogous to those of REP2; however, Dunn et al. (1994) show the two IR-A stem loops in DNA form. Top schematic depicts the DNA strand with promoter, RBS, and ATG sites on the IR-A segment.
FIG. 1.—
(a) Predicted secondary structures of repeat sequences at the RNA level. The Mfold program was used for RNA folding (Markham and Zuker 2005). The Neisseria meningitidis nemis (neisseria miniature ISs) is characteristic of MITEs, and the secondary structure shown is similar to that of Mazzone et al. (2001). The top schematic describes inverted repeats (IR) and DRs flanking the DNA strand. The bcr1 structure is that of Bacillus anthracis 1R (Økstad et al. 2004) and is typical of the Bacillus bcr1 RNA secondary structures (Klevan et al. 2007). These consist of a cruciform-like structure with two independent stem loops. The Stenotrophomonas maltophilia REP sequence and secondary structure shown is characteristic of the short high G–C content REPs found in these species; they are termed SMAG (Rocco et al. 2010). These SMAG units can carry an unpaired tetranucleotide sequence at one end. (b) Left, predicted RNA secondary structures of the REP2 sequence from N. meningitidis showing internal stem loops 1 and 2. The nt sequence is from Morelle et al. (2003). Upper schematic denotes the REP2 DNA strand with promoter, ribosome binding site (RBS), and ATG initiation codon. (b) Right, predicted secondary structural model of the Borrelia burgdoferi IR-A sequence from circular plasmid cp8.3/Ip21 [nt sequence from Dunn et al. (1994)]. Stem loops 1 and 2 may be analogous to those of REP2; however, Dunn et al. (1994) show the two IR-A stem loops in DNA form. Top schematic depicts the DNA strand with promoter, RBS, and ATG sites on the IR-A segment.
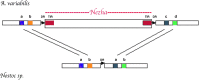
FIG. 2.—
Diagrammatic representation of empty and filled site in homologous chromosomal regions in Anabaena variabilis and Nostoc sp. (based on Zhou et al. 2008) The Nezha MITE insertion is shown in A. variabilis. Shown also diagrammatically are the DRs and TIR. Genes depicted as “a” and “b” are orthologs between the two species. In another chromosomal region (not shown), Nezha can be found inserted into a site in Nostoc sp., while the same site is empty in A. variabilis (Zhou et al. 2008).
FIG. 3.—
Diagrammatic representation of the defective integron flanked by identical IMU elements as found on Enterobacter cloacae plasmid pCHE-A (based on Poirel et al. 2009). The arrows represent the IMU inverted repeats (IR). Shown also are is the defective int1 gene at the 5′ side (left), _bla_GES-5, the beta-lactamase gene cassette in the middle, and the defective quaternary ammonium salt gene qacE on the 3′ side. Lengths are not drawn to scale.
Similar articles
- Identification of novel MITEs (miniature inverted-repeat transposable elements) in Coxiella burnetii: implications for protein and small RNA evolution.
Wachter S, Raghavan R, Wachter J, Minnick MF. Wachter S, et al. BMC Genomics. 2018 Apr 11;19(1):247. doi: 10.1186/s12864-018-4608-y. BMC Genomics. 2018. PMID: 29642859 Free PMC article. - A versatile palindromic amphipathic repeat coding sequence horizontally distributed among diverse bacterial and eucaryotic microbes.
Röske K, Foecking MF, Yooseph S, Glass JI, Calcutt MJ, Wise KS. Röske K, et al. BMC Genomics. 2010 Jul 13;11:430. doi: 10.1186/1471-2164-11-430. BMC Genomics. 2010. PMID: 20626840 Free PMC article. - Clustered regularly interspaced short palindromic repeats (CRISPRs): the hallmark of an ingenious antiviral defense mechanism in prokaryotes.
Al-Attar S, Westra ER, van der Oost J, Brouns SJ. Al-Attar S, et al. Biol Chem. 2011 Apr;392(4):277-89. doi: 10.1515/BC.2011.042. Epub 2011 Feb 7. Biol Chem. 2011. PMID: 21294681 Review. - Small mobile sequences in bacteria display diverse structure/function motifs.
Delihas N. Delihas N. Mol Microbiol. 2008 Feb;67(3):475-81. doi: 10.1111/j.1365-2958.2007.06068.x. Epub 2007 Dec 10. Mol Microbiol. 2008. PMID: 18086200 Free PMC article. Review.
Cited by
- Trichodesmium genome maintains abundant, widespread noncoding DNA in situ, despite oligotrophic lifestyle.
Walworth N, Pfreundt U, Nelson WC, Mincer T, Heidelberg JF, Fu F, Waterbury JB, Glavina del Rio T, Goodwin L, Kyrpides NC, Land ML, Woyke T, Hutchins DA, Hess WR, Webb EA. Walworth N, et al. Proc Natl Acad Sci U S A. 2015 Apr 7;112(14):4251-6. doi: 10.1073/pnas.1422332112. Epub 2015 Mar 23. Proc Natl Acad Sci U S A. 2015. PMID: 25831533 Free PMC article. - Complete sequences of two plasmids in a blaNDM-1-positive Klebsiella oxytoca isolate from Taiwan.
Huang TW, Wang JT, Lauderdale TL, Liao TL, Lai JF, Tan MC, Lin AC, Chen YT, Tsai SF, Chang SC. Huang TW, et al. Antimicrob Agents Chemother. 2013 Aug;57(8):4072-6. doi: 10.1128/AAC.02266-12. Epub 2013 Jun 10. Antimicrob Agents Chemother. 2013. PMID: 23752513 Free PMC article. - Detection and characterization of miniature inverted-repeat transposable elements in “Candidatus Liberibacter asiaticus”.
Wang X, Tan J, Bai Z, Deng X, Li Z, Zhou C, Chen J. Wang X, et al. J Bacteriol. 2013 Sep;195(17):3979-86. doi: 10.1128/JB.00413-13. J Bacteriol. 2013. PMID: 23813735 Free PMC article. - How Do Transposable Elements Activate Expression of Transcriptionally Silent Antibiotic Resistance Genes?
Lipszyc A, Szuplewska M, Bartosik D. Lipszyc A, et al. Int J Mol Sci. 2022 Jul 22;23(15):8063. doi: 10.3390/ijms23158063. Int J Mol Sci. 2022. PMID: 35897639 Free PMC article. Review. - Long-read sequencing for identification of insertion sites in large transposon mutant libraries.
Yasir M, Turner AK, Lott M, Rudder S, Baker D, Bastkowski S, Page AJ, Webber MA, Charles IG. Yasir M, et al. Sci Rep. 2022 Mar 3;12(1):3546. doi: 10.1038/s41598-022-07557-x. Sci Rep. 2022. PMID: 35241765 Free PMC article.
References
- Al-Attar S, Westra ER, van der Oost J, Brouns SJ. Clustered regularly interspaced short palindromic repeats (CRISPRs): the hallmark of an ingenious antiviral defense mechanism in prokaryotes. Biol Chem. 2011;392:277–289. - PubMed
- Bachellier S, Clément JM, Hofnung M. Short palindromic repetitive DNA elements in enterobacteria: a survey. Res Microbiol. 1999;150:627–639. - PubMed
- Bachellier S, Saurin W, Perrin D, Hofnung M, Gilson E. Structural and functional diversity among bacterial interspersed mosaic elements (BIMEs) Mol Microbiol. 1994;12:61–70. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous