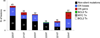Analysis of the coding genome of diffuse large B-cell lymphoma - PubMed (original) (raw)
. 2011 Jul 31;43(9):830-7.
doi: 10.1038/ng.892.
Vladimir Trifonov, Giulia Fabbri, Jing Ma, Davide Rossi, Annalisa Chiarenza, Victoria A Wells, Adina Grunn, Monica Messina, Oliver Elliot, Joseph Chan, Govind Bhagat, Amy Chadburn, Gianluca Gaidano, Charles G Mullighan, Raul Rabadan, Riccardo Dalla-Favera
Affiliations
- PMID: 21804550
- PMCID: PMC3297422
- DOI: 10.1038/ng.892
Analysis of the coding genome of diffuse large B-cell lymphoma
Laura Pasqualucci et al. Nat Genet. 2011.
Abstract
Diffuse large B-cell lymphoma (DLBCL) is the most common form of human lymphoma. Although a number of structural alterations have been associated with the pathogenesis of this malignancy, the full spectrum of genetic lesions that are present in the DLBCL genome, and therefore the identity of dysregulated cellular pathways, remains unknown. By combining next-generation sequencing and copy number analysis, we show that the DLBCL coding genome contains, on average, more than 30 clonally represented gene alterations per case. This analysis also revealed mutations in genes not previously implicated in DLBCL pathogenesis, including those regulating chromatin methylation (MLL2; 24% of samples) and immune recognition by T cells. These results provide initial data on the complexity of the DLBCL coding genome and identify novel dysregulated pathways underlying its pathogenesis.
Figures
Figure 1. DLBCL non-silent mutation load
Number (a) and type (b) of Sanger-validated, non-silent mutations identified in the 6 DLBCL discovery patients. c, Nucleotide targeting of the DLBCL-associated point mutations (nucleotide composition of the target exome: G, 26.2%; C, 25.7%; T, 22.0%; A, 25.9%). d, Observed mutation frequencies at specific dinucleotides (red bars). The expected frequencies (grey bars) correpond to the dinucleotide sequence composition of the target exome. Asterisks denote statistically significant differences in overrepresented changes, as assessed by a Poisson distribution after correction for multiple hypotheses.
Figure 2. Copy number analysis of the 6 DLBCL discovery cases
a, dChipSNP heatmap showing median-smoothed log2 copy number ratio of all chromosomes in the 6 index DLBCL biopsies and their paired normal DNAs. In the red-blue scale, white corresponds to a normal (diploid) copy number log-ratio, blue is deletion and red is gain. b, Curated segmentation data for the 6 DLBCL samples shown in panel a, obtained as described in Methods and visualized using the Integrative Genomics Viewer (IGV) software (
http://www.broadinstitute.org/igv
). b, Overall number and frequency of copy number alterations.
Figure 3. DLBCL harbors a heterogeneous load of numerical and structural genomic aberrations
Combined load of genetic lesions that were present in the major tumor clone of the 6 DLBCL discovery cases (including point mutations, copy number aberrations, and chromosomal translocations at three common target loci).
Figure 4. Recurrent mutations in DLBCL
Percentage of DLBCL primary cases harboring mutations in 56 candidate genes after targeted-resequencing of an expanded screening dataset. The final number of mutated cases over total analyzed (including the 6 DLBCL discovery cases) is given for each gene; this number includes validated somatic mutations as well as variants that are not reported in any available SNP databases (see Methods). The somatic origin of the mutations was confirmed by analysis of paired normal DNA in at least one case/gene. Red asterisks denote putative tumor suppressor genes, as suggested by the presence of predominantly inactivating mutations (stop codons, frameshift mutations and experimentally validated missense mutations). Green asterisks denote genes where the oncogenic activity of the mutation was functionally proven. Black asterisks indicate genes harboring mutations of unclear pathogenic role, but predicted to truncate the encoded protein in at least one case.
Figure 5. The MLL2 gene is mutated in a large fraction of DLBCL
a, Schematic diagram of the MLL2 gene (top) and protein (bottom), with its conserved functional domains (PHD, FYRN, FYRC, SET). In black, untranslated exons. Arrows, color-coded as in (b), indicate the position of the mutations found (cell lines on top, primary cases at the bottom). b, Overall percentage of DLBCL cases carrying MLL2 mutations, according to mutation type (cell lines and biopsies). c, Prevalence of MLL2 mutated cases in major DLBCL phenotypic subtypes; numbers on top indicate the actual number of mutated samples over total analyzed. d, Allelic distribution of the mutations in 26 of 28 affected samples for which this data could be obtained. Mutations are color-coded as in (b).
Figure 6. Disruption of histone/chromatin modification genes is a major feature of DLBCL
a, Mutation frequency of five genes encoding for histone-acetyltransferases and histone-methylation modifiers in GCB- and ABC-DLBCL (primary cases only). b, dChipSNP heatmap showing median-smoothed log2 copy number ratio for DLBCL biopsies harboring KDM2B, MLL3 and MLL5 deletions; in the red-blue scale, white corresponds to a normal (diploid) copy number, blue is deletion and red is gain. Note that, due to the presence of non-tumor cells infiltrating the biopsies, the inferred copy number, and the corresponding color intensity, may vary across samples (see Supplementary Table 11). The boxed area corresponds to the commonly deleted region, which is expanded below the heatmap to show the included genes. c, Relationship between MLL2 mutations and mutations of HATs. In the heatmap, rows correspond to the indicated genes and columns represent individual samples, color-coded according to the gene status (grey, unmutated; red, mutated or deleted). Of the 3 _MLL2_-mutated cases showing simultaneous lesions at CREBBP, one harbored an amino acid substitution whose functional significance is presently unclear. Thus, it is possible that this change represents a passenger event or a private germline variant not annotated in currently available databases; alternatively, these mutations may have milder effects, requiring additional cooperating lesions in the same pathway. d, Overall fraction of DLBCL biopsies carrying mutations at one or more of the four histone modification genes shown in (c).
Similar articles
- Prediction and characterization of diffuse large B-cell lymphoma cell-of-origin subtypes using targeted sequencing.
Trabucco SE, Sokol ES, Maund SL, Moore JA, Frampton GM, Albacker LA, Oestergaard MZ, Venstrom J, Sehn LH, Bolen CR. Trabucco SE, et al. Future Oncol. 2021 Nov;17(31):4171-4183. doi: 10.2217/fon-2021-0370. Epub 2021 Jul 27. Future Oncol. 2021. PMID: 34313135 - Characterization of genomic imbalances in diffuse large B-cell lymphoma by detailed SNP-chip analysis.
Scholtysik R, Kreuz M, Hummel M, Rosolowski M, Szczepanowski M, Klapper W, Loeffler M, Trümper L, Siebert R, Küppers R; Molecular Mechanisms in Malignant Lymphomas Network Project of the Deutsche Krebshilfe. Scholtysik R, et al. Int J Cancer. 2015 Mar 1;136(5):1033-42. doi: 10.1002/ijc.29072. Epub 2014 Jul 22. Int J Cancer. 2015. PMID: 25042405 - Mutational and structural analysis of diffuse large B-cell lymphoma using whole-genome sequencing.
Morin RD, Mungall K, Pleasance E, Mungall AJ, Goya R, Huff RD, Scott DW, Ding J, Roth A, Chiu R, Corbett RD, Chan FC, Mendez-Lago M, Trinh DL, Bolger-Munro M, Taylor G, Hadj Khodabakhshi A, Ben-Neriah S, Pon J, Meissner B, Woolcock B, Farnoud N, Rogic S, Lim EL, Johnson NA, Shah S, Jones S, Steidl C, Holt R, Birol I, Moore R, Connors JM, Gascoyne RD, Marra MA. Morin RD, et al. Blood. 2013 Aug 15;122(7):1256-65. doi: 10.1182/blood-2013-02-483727. Epub 2013 May 22. Blood. 2013. PMID: 23699601 Free PMC article. - [Analysis of genomic copy number alterations of malignant lymphomas and its application for diagnosis].
Tagawa H. Tagawa H. Gan To Kagaku Ryoho. 2007 Jul;34(7):975-82. Gan To Kagaku Ryoho. 2007. PMID: 17637530 Review. Japanese. - Dissecting aggressive B-cell lymphoma through genomic analysis - What is clinically relevant?
Scott DW, Rimsza LM. Scott DW, et al. Best Pract Res Clin Haematol. 2018 Sep;31(3):187-198. doi: 10.1016/j.beha.2018.07.003. Epub 2018 Jul 7. Best Pract Res Clin Haematol. 2018. PMID: 30213388 Review.
Cited by
- Specific Mutation Predict Relapse/Refractory Diffuse Large B-Cell Lymphoma.
Wang J, Tian L, Zhang W, Tang S, Zhao W, Guo Y, Wu C, Lin Y, Ke X, Jing H. Wang J, et al. J Blood Med. 2024 Sep 10;15:407-419. doi: 10.2147/JBM.S471639. eCollection 2024. J Blood Med. 2024. PMID: 39279878 Free PMC article. - Development of New Diffuse Large B Cell Lymphoma Mouse Models.
Mehdi SH, Xu YZ, Shultz LD, Kim E, Lee YG, Kendrick S, Yoon D. Mehdi SH, et al. Cancers (Basel). 2024 Aug 29;16(17):3006. doi: 10.3390/cancers16173006. Cancers (Basel). 2024. PMID: 39272864 Free PMC article. - SETD1B mutations confer apoptosis resistance and BCL2 independence in B cell lymphoma.
Portelinha A, Wang S, Parsa S, Jiang M, Gorelick AN, Mohanty S, Sharma S, de Stanchina E, Berishaj M, Zhao C, Heward J, Aryal NK, Tavana O, Wen J, Fitzgibbon J, Dogan A, Younes A, Melnick AM, Wendel HG. Portelinha A, et al. J Exp Med. 2024 Oct 7;221(10):e20231143. doi: 10.1084/jem.20231143. Epub 2024 Sep 5. J Exp Med. 2024. PMID: 39235528 - MEF2B C-terminal mutations enhance transcriptional activity and stability to drive B cell lymphomagenesis.
Yu C, Shen Q, Holmes AB, Mo T, Tosato A, Soni RK, Corinaldesi C, Koul S, Pasqualucci L, Hussein S, Forouhar F, Dalla-Favera R, Basso K. Yu C, et al. Nat Commun. 2024 Aug 21;15(1):7195. doi: 10.1038/s41467-024-51644-8. Nat Commun. 2024. PMID: 39179580 Free PMC article. - Silencing GNAS enhances HDAC3i efficacy in CREBBP wild type B cell lymphoma.
Mondello P. Mondello P. Leukemia. 2024 Oct;38(10):2087-2089. doi: 10.1038/s41375-024-02355-y. Epub 2024 Jul 19. Leukemia. 2024. PMID: 39030358 Review.
References
- Abramson JS, Shipp MA. Advances in the biology and therapy of diffuse large B-cell lymphoma: moving toward a molecularly targeted approach. Blood. 2005;106:1164–1174. - PubMed
- Swerdlow SH, et al. Lyon: International Agency for Research on Cancer (IARC); 2008. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues.
- Alizadeh AA, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. - PubMed
- Savage KJ, et al. The molecular signature of mediastinal large B-cell lymphoma differs from that of other diffuse large B-cell lymphomas and shares features with classical Hodgkin lymphoma. Blood. 2003;102:3871–3879. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U54-AI057158/AI/NIAID NIH HHS/United States
- R01-CA37295/CA/NCI NIH HHS/United States
- R01 CA037295/CA/NCI NIH HHS/United States
- P01 CA092625/CA/NCI NIH HHS/United States
- U54 CA121852/CA/NCI NIH HHS/United States
- U54 AI057158/AI/NIAID NIH HHS/United States
- P01 CA092625-10/CA/NCI NIH HHS/United States
- P01-CA092625/CA/NCI NIH HHS/United States
- 1R01LM010140-01/LM/NLM NIH HHS/United States
- CA121852-05/CA/NCI NIH HHS/United States
- R01 CA037295-20/CA/NCI NIH HHS/United States
- R01 LM010140/LM/NLM NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous