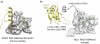Handpicking epigenetic marks with PHD fingers - PubMed (original) (raw)
Review
. 2011 Nov;39(21):9061-71.
doi: 10.1093/nar/gkr613. Epub 2011 Aug 3.
Affiliations
- PMID: 21813457
- PMCID: PMC3241642
- DOI: 10.1093/nar/gkr613
Review
Handpicking epigenetic marks with PHD fingers
Catherine A Musselman et al. Nucleic Acids Res. 2011 Nov.
Abstract
Plant homeodomain (PHD) fingers have emerged as one of the largest families of epigenetic effectors capable of recognizing or 'reading' post-translational histone modifications and unmodified histone tails. These interactions are highly specific and can be modulated by the neighboring epigenetic marks and adjacent effectors. A few PHD fingers have recently been found to also associate with non-histone proteins. In this review, we detail the molecular mechanisms and biological outcomes of the histone and non-histone targeting by PHD fingers. We discuss the significance of crosstalk between the histone modifications and consequences of combinatorial readout for selective recruitment of the PHD finger-containing components of chromatin remodeling and transcriptional complexes.
Figures
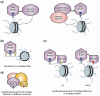
Figure 1.
PHD fingers as epigenetic effectors. (a) Histone-recognizing PHD fingers are commonly found in enzymes (left) and proteins that stabilize enzymatic complexes at chromatin (right) to further modify DNA and histones. (b–d) The specificity of a PHD finger can be increased by (b) sensitivity to multiple PTMs, (c) combinatorial readout by multiple effectors in the same protein and (d) combinatorial action of multiple effectors in different subunits of a complex. The effectors could recognize PTMs on a single histone tail (cis mechanism) or different histone tails (trans mechanism).
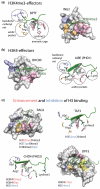
Figure 2.
The molecular mechanism of histone recognition by the PHD fingers. PHD fingers are specific for (a) H3K4me3 or (b) unmodified H3K4. The histone-binding sites of the BPTF (2F6J), ING2 (2G6Q), BHC80 (2PUY) and AIRE (2KE1) PHD fingers are shown. The binding pockets for Ala1, Arg2, Lys4me3 (or Lys4) and Lys9 of the H3K4me3 and H3K4 peptides are colored light blue, orange, pink and light green, respectively. The bound peptides are shown as a ribbon diagram and colored green. (c) Binding of the PHD fingers to H3K4me3 and H3K4 is modulated by additional PTMs. The structures of the PHD fingers of RAG2 (2V87), TAF3 (2K17), CHD4 (2L75) and DPF3 (2KWJ) are colored as in (a and b). PTMs that enhance or inhibit recognition of the primary PTM are listed and colored red and blue, respectively. An aspartate residue in the aromatic cage of TAF3 and the Lys14ac-binding pocket of DPF3 are colored wheat and yellow, respectively.
Figure 3.
The structural basis of non-histone recognition by PHD fingers. (a) The ternary complex of the PHD finger of PYGO1 (2VPG). (b) The PHD finger of MLL1 binds to H3K4me3 (3LQJ) and the RRM domain of Cyp33 (2KU7).
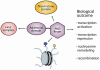
Figure 4.
The biological outcome of the recognition of histone or non-histone proteins by a PHD finger depends on the function of the complex in which the PHD finger resides and the local regulatory environment.
Similar articles
- Structural insight into histone recognition by the ING PHD fingers.
Champagne KS, Kutateladze TG. Champagne KS, et al. Curr Drug Targets. 2009 May;10(5):432-41. doi: 10.2174/138945009788185040. Curr Drug Targets. 2009. PMID: 19442115 Free PMC article. Review. - Characterization of the plant homeodomain (PHD) reader family for their histone tail interactions.
Jain K, Fraser CS, Marunde MR, Parker MM, Sagum C, Burg JM, Hall N, Popova IK, Rodriguez KL, Vaidya A, Krajewski K, Keogh MC, Bedford MT, Strahl BD. Jain K, et al. Epigenetics Chromatin. 2020 Jan 24;13(1):3. doi: 10.1186/s13072-020-0328-z. Epigenetics Chromatin. 2020. PMID: 31980037 Free PMC article. - Molecular investigation of the tandem Tudor domain and plant homeodomain histone binding domains of the epigenetic regulator UHRF2.
Ginnard SM, Winkler AE, Mellado Fritz C, Bluhm T, Kemmer R, Gilliam M, Butkevich N, Abdrabbo S, Bricker K, Feiler J, Miller I, Zoerman J, El-Mohri Z, Khuansanguan P, Basch M, Petzold T, Kostoff M, Konopka S, Kociba B, Gillis T, Heyl DL, Trievel RC, Albaugh BN. Ginnard SM, et al. Proteins. 2022 Mar;90(3):835-847. doi: 10.1002/prot.26278. Epub 2021 Nov 19. Proteins. 2022. PMID: 34766381 - Multivalent recognition of histone tails by the PHD fingers of CHD5.
Oliver SS, Musselman CA, Srinivasan R, Svaren JP, Kutateladze TG, Denu JM. Oliver SS, et al. Biochemistry. 2012 Aug 21;51(33):6534-44. doi: 10.1021/bi3006972. Epub 2012 Aug 8. Biochemistry. 2012. PMID: 22834704 Free PMC article. - PHD fingers: epigenetic effectors and potential drug targets.
Musselman CA, Kutateladze TG. Musselman CA, et al. Mol Interv. 2009 Dec;9(6):314-23. doi: 10.1124/mi.9.6.7. Mol Interv. 2009. PMID: 20048137 Free PMC article. Review.
Cited by
- The BNB-GLID module regulates germline fate determination in Marchantia polymorpha.
Ren X, Zhang X, Qi X, Zhang T, Wang H, Twell D, Gong Y, Fu Y, Wang B, Kong H, Xu B. Ren X, et al. Plant Cell. 2024 Sep 3;36(9):3824-3837. doi: 10.1093/plcell/koae206. Plant Cell. 2024. PMID: 39041486 Free PMC article. - Intestinal NSD2 Aggravates Nonalcoholic Steatohepatitis Through Histone Modifications.
Zhang Y, Qiao Y, Li Z, Liu D, Jin Q, Guo J, Li X, Chen L, Liu L, Peng L. Zhang Y, et al. Adv Sci (Weinh). 2024 Sep;11(33):e2402551. doi: 10.1002/advs.202402551. Epub 2024 Jun 26. Adv Sci (Weinh). 2024. PMID: 38923875 Free PMC article. - Genome-Wide Identification and Expression Analysis of the PHD Finger Gene Family in Pea (Pisum sativum).
Liu M, Li W, Zheng X, Yuan Z, Zhou Y, Yang J, Mao Y, Wang D, Wu Q, He Y, He L, Zong D, Chen J. Liu M, et al. Plants (Basel). 2024 May 28;13(11):1489. doi: 10.3390/plants13111489. Plants (Basel). 2024. PMID: 38891298 Free PMC article. - Genome-wide identification of the plant homeodomain-finger family in rye and ScPHD5 functions in cold tolerance and flowering time.
Jung WJ, Jeong JH, Yoon JS, Seo YW. Jung WJ, et al. Plant Cell Rep. 2024 May 15;43(6):142. doi: 10.1007/s00299-024-03226-7. Plant Cell Rep. 2024. PMID: 38744747 - Bromodomain inhibition targeting BPTF in the treatment of melanoma and other solid tumors.
Khan I, Kashani-Sabet M. Khan I, et al. Clin Exp Metastasis. 2024 Aug;41(4):509-515. doi: 10.1007/s10585-024-10265-7. Epub 2024 Apr 29. Clin Exp Metastasis. 2024. PMID: 38683257 Review.
References
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. - PubMed
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. - PubMed
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. - PubMed
- Turner BM. Cellular memory and the histone code. Cell. 2002;111:285–291. - PubMed
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. - PubMed