Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA - PubMed (original) (raw)
. 2011 Sep 2;333(6047):1303-7.
doi: 10.1126/science.1210944. Epub 2011 Aug 4.
Bin-Zhong Li, Zheng Li, Peng Liu, Yang Wang, Qingyu Tang, Jianping Ding, Yingying Jia, Zhangcheng Chen, Lin Li, Yan Sun, Xiuxue Li, Qing Dai, Chun-Xiao Song, Kangling Zhang, Chuan He, Guo-Liang Xu
Affiliations
- PMID: 21817016
- PMCID: PMC3462231
- DOI: 10.1126/science.1210944
Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA
Yu-Fei He et al. Science. 2011.
Abstract
The prevalent DNA modification in higher organisms is the methylation of cytosine to 5-methylcytosine (5mC), which is partially converted to 5-hydroxymethylcytosine (5hmC) by the Tet (ten eleven translocation) family of dioxygenases. Despite their importance in epigenetic regulation, it is unclear how these cytosine modifications are reversed. Here, we demonstrate that 5mC and 5hmC in DNA are oxidized to 5-carboxylcytosine (5caC) by Tet dioxygenases in vitro and in cultured cells. 5caC is specifically recognized and excised by thymine-DNA glycosylase (TDG). Depletion of TDG in mouse embyronic stem cells leads to accumulation of 5caC to a readily detectable level. These data suggest that oxidation of 5mC by Tet proteins followed by TDG-mediated base excision of 5caC constitutes a pathway for active DNA demethylation.
Figures
Fig. 1. Purified Tet2 catalyzes the modification of 5mC and 5hmC
(A) The 32P spot X on a TLC plate generated from 5mC and 5hmC DNA substrates incubated with the full-length Flag-Tet2 protein. The top spot in lanes 3 to 8 was 5′ end-labeled deoxyadenosine monophosphate (dAMP) resulting from the incomplete EcoNI digestion of the DNA substrate (fig. S1A). (B) TLC confirmation of the origin of spot X from 5mC with a 14C-labeled methyl group. (C) HPLC detection of a new nucleoside generated from 5mC and 5hmC DNA substrates upon incubation with Flag-Tet2. AU indicates absorption units.
Fig. 2. The modification product of 5mC is 5caC
The 5mC derivative generated by Flag-Tet2 was analyzed by HPLC analysis (A), TLC (B), and mass spectrometry (C) using a 5caC standard as a reference. (A) HPLC analysis of the nucleoside derived from 5mC in the DNA substrate treated with Flag-Tet2. (B) TLC identification of the modification product of Tet2. The 5mC substrate used was the same as in Fig. 1A. 5caC DNA (lane 3), a synthetic oligonucleotide duplex with the same sequence but containing a 5-caC 3′ of the cleavage site of EcoNI, provides a reference spot for 5-carboxycytidine monophosphate (5ca-dCMP). (C) Mass spectrometry analysis of a HPLC fraction corresponding to the peak X′ in (A). Structural formulas deduced are shown for the major peaks.
Fig. 3. Tet2 catalyzes formation of 5caC in genomic DNA in vivo
(A) HPLC detection of 5caC in genomic DNA of HEK 293T cells expressing wild-type Flag-Tet2. Synthetic 2′-deoxy-5-carboxylcytidine (5caC) and the nucleoside hydrolysate of a synthetic DNA containing cytosine (C), 5hmC, and 5mC were used as standards. Arrows point to the 5caC peaks detected in DNA isolated from cells transfected with wild-type Tet2. (B) HPLC–tandem mass spectrometry (MS/MS) detection of genomic 5caC. Shown are MRM elution profiles of negative-ion mass transitions from a precursor to its three product ions as shown in Fig. 2C for a synthetic 2′-deoxy-5-carboxycytidine standard (blue) and a DNA hydrolysate [isolated peak indicated with a red arrow in (A)] from cells transfected with full-length Tet2 (black). Red line was from a control DNA sample isolated from cells transfected with the inactive Tet2 mutant.
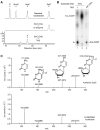
Fig. 4. TDG glycosylase recognizes and excises 5caC from DNA
(A) ES cell nuclear extract contains 5caC-specific base-excision activity. The activity in the nuclear extract of mouse ES cells to generate alkaline-sensitive sites was assayed by using 5caC-containing oligonucleotide duplexes. Shown are the results obtained with 20-mer DNA duplexes containing either G/U (U), G/5hmC (5hmC), or G/5caC (5caC) base pairs in the middle. (B) Excision of 5caC from DNA by Flag-TDG but not by Flag-MBD4, Flag-UNG, or GST-SMUG1. Asn151→Ala151 (N151A) is a catalytically inactive mutant of TDG. Proteins used are shown in fig. S10B. 6xHis-TDG purified from bacteria was also active in 5caC excision. TDG displayed a much stronger glycosylase activity for the “hemi-carboxylated” DNA substrate containing 5caC only on one strand (fig. S11). WT, wild type. (C) Reduced 5caC formation by cotransfection of TDG in HEK 293T cells expressing ectopic Tet2. The mutant TDG was as in (B). (D) Lack of 5caC base-excision activity in Tdg knockdown ES cells. Nuclear extracts prepared from the two independent cell lines (a and b) containing shRNA knockdown construct 1, 2, or scramble control were tested as in (A). (E) HPLC-MS/MS detection of 5caC in ES cells depleted of TDG. MRM profiles of hydrolysates of genomic DNA from control (red) and TDG-depleted ES cells (pink) were analyzed. Synthetic 5caC nucleoside was used as a positive control (blue). Depletion of TDG was confirmed by Western analysis of independent stable knockdown ES cell lines (fig. S12).
Comment in
- Molecular biology. Demystifying DNA demethylation.
Nabel CS, Kohli RM. Nabel CS, et al. Science. 2011 Sep 2;333(6047):1229-30. doi: 10.1126/science.1211917. Science. 2011. PMID: 21885763 No abstract available.
Similar articles
- Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine.
Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Ito S, et al. Science. 2011 Sep 2;333(6047):1300-3. doi: 10.1126/science.1210597. Epub 2011 Jul 21. Science. 2011. PMID: 21778364 Free PMC article. - Genome-wide analysis reveals TET- and TDG-dependent 5-methylcytosine oxidation dynamics.
Shen L, Wu H, Diep D, Yamaguchi S, D'Alessio AC, Fung HL, Zhang K, Zhang Y. Shen L, et al. Cell. 2013 Apr 25;153(3):692-706. doi: 10.1016/j.cell.2013.04.002. Epub 2013 Apr 18. Cell. 2013. PMID: 23602152 Free PMC article. - Roles of TET and TDG in DNA demethylation in proliferating and non-proliferating immune cells.
Onodera A, González-Avalos E, Lio CJ, Georges RO, Bellacosa A, Nakayama T, Rao A. Onodera A, et al. Genome Biol. 2021 Jun 22;22(1):186. doi: 10.1186/s13059-021-02384-1. Genome Biol. 2021. PMID: 34158086 Free PMC article. - Epigenetic modifications in DNA could mimic oxidative DNA damage: A double-edged sword.
Ito S, Kuraoka I. Ito S, et al. DNA Repair (Amst). 2015 Aug;32:52-57. doi: 10.1016/j.dnarep.2015.04.013. Epub 2015 May 1. DNA Repair (Amst). 2015. PMID: 25956859 Review. - MicroRNAs mediated targeting on the Yin-yang dynamics of DNA methylation in disease and development.
Tu J, Liao J, Luk AC, Tang NL, Chan WY, Lee TL. Tu J, et al. Int J Biochem Cell Biol. 2015 Oct;67:115-20. doi: 10.1016/j.biocel.2015.05.002. Epub 2015 May 12. Int J Biochem Cell Biol. 2015. PMID: 25979370 Review.
Cited by
- Epigenetics of neural differentiation: Spotlight on enhancers.
Giacoman-Lozano M, Meléndez-Ramírez C, Martinez-Ledesma E, Cuevas-Diaz Duran R, Velasco I. Giacoman-Lozano M, et al. Front Cell Dev Biol. 2022 Oct 13;10:1001701. doi: 10.3389/fcell.2022.1001701. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36313573 Free PMC article. Review. - Acute deletion of TET enzymes results in aneuploidy in mouse embryonic stem cells through decreased expression of Khdc3.
Georges RO, Sepulveda H, Angel JC, Johnson E, Palomino S, Nowak RB, Desai A, López-Moyado IF, Rao A. Georges RO, et al. Nat Commun. 2022 Oct 20;13(1):6230. doi: 10.1038/s41467-022-33742-7. Nat Commun. 2022. PMID: 36266342 Free PMC article. - The histone deacetylase SIRT6 controls embryonic stem cell fate via TET-mediated production of 5-hydroxymethylcytosine.
Etchegaray JP, Chavez L, Huang Y, Ross KN, Choi J, Martinez-Pastor B, Walsh RM, Sommer CA, Lienhard M, Gladden A, Kugel S, Silberman DM, Ramaswamy S, Mostoslavsky G, Hochedlinger K, Goren A, Rao A, Mostoslavsky R. Etchegaray JP, et al. Nat Cell Biol. 2015 May;17(5):545-57. doi: 10.1038/ncb3147. Epub 2015 Apr 27. Nat Cell Biol. 2015. PMID: 25915124 Free PMC article. - Identification of CD34+ and CD34- leukemia-initiating cells in MLL-rearranged human acute lymphoblastic leukemia.
Aoki Y, Watanabe T, Saito Y, Kuroki Y, Hijikata A, Takagi M, Tomizawa D, Eguchi M, Eguchi-Ishimae M, Kaneko A, Ono R, Sato K, Suzuki N, Fujiki S, Koh K, Ishii E, Shultz LD, Ohara O, Mizutani S, Ishikawa F. Aoki Y, et al. Blood. 2015 Feb 5;125(6):967-80. doi: 10.1182/blood-2014-03-563304. Epub 2014 Dec 23. Blood. 2015. PMID: 25538041 Free PMC article. - The Influence of Hydroxylation on Maintaining CpG Methylation Patterns: A Hidden Markov Model Approach.
Giehr P, Kyriakopoulos C, Ficz G, Wolf V, Walter J. Giehr P, et al. PLoS Comput Biol. 2016 May 25;12(5):e1004905. doi: 10.1371/journal.pcbi.1004905. eCollection 2016 May. PLoS Comput Biol. 2016. PMID: 27224554 Free PMC article.
References
- Jaenisch R, Bird A. Nat Genet. 2003;33(suppl):245. - PubMed
- Simonsson S, Gurdon J. Nat Cell Biol. 2004;6:984. - PubMed
- Liutkeviciute Z, Lukinavicius G, Masevicius V, Daujotyte D, Klimasauskas S. Nat Chem Biol. 2009;5:400. - PubMed
- Walsh CP, Xu GL. Curr Top Microbiol Immunol. 2006;301:283. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM071440/GM/NIGMS NIH HHS/United States
- S10 RR027643/RR/NCRR NIH HHS/United States
- GM071440/GM/NIGMS NIH HHS/United States
- 1S10RR027643-01/RR/NCRR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases