Direct targeting of Sec23a by miR-200s influences cancer cell secretome and promotes metastatic colonization - PubMed (original) (raw)
. 2011 Aug 7;17(9):1101-8.
doi: 10.1038/nm.2401.
Brian J Ell, Francesca M Buffa, Toni Ibrahim, Mario A Blanco, Toni Celià-Terrassa, Laura Mercatali, Zia Khan, Hani Goodarzi, Yuling Hua, Yong Wei, Guohong Hu, Benjamin A Garcia, Jiannis Ragoussis, Dino Amadori, Adrian L Harris, Yibin Kang
Affiliations
- PMID: 21822286
- PMCID: PMC3169707
- DOI: 10.1038/nm.2401
Direct targeting of Sec23a by miR-200s influences cancer cell secretome and promotes metastatic colonization
Manav Korpal et al. Nat Med. 2011.
Abstract
Although the role of miR-200s in regulating E-cadherin expression and epithelial-to-mesenchymal transition is well established, their influence on metastatic colonization remains controversial. Here we have used clinical and experimental models of breast cancer metastasis to discover a pro-metastatic role of miR-200s that goes beyond their regulation of E-cadherin and epithelial phenotype. Overexpression of miR-200s is associated with increased risk of metastasis in breast cancer and promotes metastatic colonization in mouse models, phenotypes that cannot be recapitulated by E-cadherin expression alone. Genomic and proteomic analyses revealed global shifts in gene expression upon miR-200 overexpression toward that of highly metastatic cells. miR-200s promote metastatic colonization partly through direct targeting of Sec23a, which mediates secretion of metastasis-suppressive proteins, including Igfbp4 and Tinagl1, as validated by functional and clinical correlation studies. Overall, these findings suggest a pleiotropic role of miR-200s in promoting metastatic colonization by influencing E-cadherin-dependent epithelial traits and Sec23a-mediated tumor cell secretome.
Conflict of interest statement
COMPETING INTERESTS STATEMENT The authors declare that they have no competing financial interests.
Figures
Figure 1
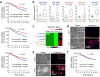
MiR-200s are associated with poor prognosis in breast cancer. (a) Kaplan-Meier curves showing the distant relapse-free survival of 210 patients with high or low expression of the entire miR-200 family (top panel), miR-429 (middle panel) and miR-200a (bottom panel) in breast tumors. P values were computed by a likelihood ratio test. (b) Box plots showing miR-200 expression levels in ten human primary and metastasis samples as assessed by qRT-PCR analysis. Data is normalized to U6 and P values were computed by Student’s t-test. (c) Heat map showing miRNA expression levels in 4T1 series. 168: 168FARN. (d) Phase contrast images (left panel) and immunofluorescence (IF) images of 4TO7 and 4T1 cells stained for E-cadherin (right panel). (e) Phase contrast images (left panel) and IF images for E-cadherin (right panel) of MCFCA1h and MCFCA1a cells. Inserts highlight the membrane localization of E-cadherin. (f) Kaplan-Meier curves showing the distant-relapse free survival of 210 patients with high or low CDH1 expression. P values were computed by a likelihood ratio test.
Figure 2
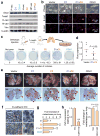
Ectopic miR-200 expression enhances spontaneous metastasis and colonization of distant organs. (a) Western blot showing expression of indicated proteins in various genetically modified 4TO7 cell lines. (b) Phase contrast and IF images of cell lines stained for E-cadherin and N-cadherin. Yellow outline emphasizes cell morphology. (c) Various cell lines were used to generate orthotopic mammary gland tumors. After one month, lungs were excised, dissociated and plated in selective media for colony formation. Average numbers of colonies were listed below representative plate images. Data represent mean ± s.e.m. from a single representative experiment out of three independent experiments. (n = 9–10). (d) Dot plot showing relative expression of puromycin resistance gene, an indicator of circulating tumor cells, by qRT-PCR analysis of genomic DNA from whole blood samples. Red lines represent median values. P = 0.02 (Student’s t-test). (e) Representative gross lung and H&E stained lung sections from animals intravenously injected with various 4TO7 cell lines. Red arrowheads and dotted lines highlight metastatic nodules. (f) Immunohistochemical staining for E-cadherin of lung nodules established from indicated cells. (g) Bar graph showing fold increase in number of pulmonary metastasis nodules for each group. Data represent mean fold increase ± s.e.m. from a single representative experiment out of three independent experiments. (n = 9–10). (h) Left: RT-PCR showing expression of Cdh1 in C1C2 cells with or without stable _Cdh1_knockdown. Right: Bar graph showing number of pulmonary lesions following intravenous inoculation of tumor cells. * P < 0.05, ** P < 0.01 (Student’s t-test).
Figure 3
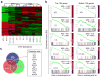
Ectopic miR-200 expression promotes global changes in gene expression. (a) Unsupervised clustering highlighting genome-wide changes in gene expression upon miR-200 expression in 4TO7 cells. Experiment was performed twice in duplicates. (b) GSEA showing influence of miR-200 overexpression on the overall gene expression profile of 4TO7 cells. Gene sets used are the top 100 (left panel) and bottom 100 (right panel) differentially expressed genes in the test (C1, C2, C1+C2 and CDH1) vs control lines. Gene list used include all mouse genes ranked by their differential expression between 4T1 and 4TO7. Enrichment of top and bottom 100 genes from 4T1 vs. 4TO7 ranked list is shown as an example of maximum possible enrichment. ES: enrichment score. NES: normalized enrichment score. Red and blue arrows denote the relative number of core genes for each analysis. (c) Venn diagram showing significant overlap of core genes from top 100 gene sets for C1 (red circle), C2 (blue circle) and C1+C2 (green circle) lines from (b). Common core genes between C1, C2 and C1+C2 lines are presented. Cdh1 is highlighted in red font to emphasize the positive influence of miR-200s on E-cadherin expression.
Figure 4
Identification of putative miR-200 targets using mass spectrometry. (a) Scatter plot shows the comparison between protein abundance and mRNA abundance for 1562 proteins in C1+C2 versus control cells. Red dots represent genes with miR-200 target sites. (b) Comparison between protein abundance and mRNA abundance for genes containing miR-200 target sites (n =130). Genes highlighted by red dots represent those that are significantly reduced in expression at both mRNA and protein level (n = 9). Dots confined within orange dashed circle represent genes harboring miR-200 target sites showing little or no change in gene expression. (c) qRT-PCR validation of reduced expression of the nine candidate genes highlighted by red dots in panel (b) in C1+C2 line compared to the control. Data represent mean ± s.e.m. * P < 0.05 (Student’s t-test). (d) Heat map showing expression of nine candidate genes in MDA-MB-231 (left) and TSU-PR1 (right) cells upon transient transfection of miR-200s. ZEB1 and ZEB2 were included as positive controls. (e) Heat map showing negative correlation between average expression of nine-candidate gene signature and miRs-200b and -200c in NCI-60 panel of cell lines. (f) Direct targeting of eight of nine candidate genes was tested by luciferase assays in HeLa cells. Data represents percent change in normalized luciferase activity after co-transfection of miR-200s relative to the negative control pre-miR ± s.e.m. * P < 0.05, ** P < 0.01 (Student’s t-test).
Figure 5
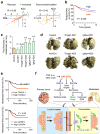
Sec23a knockdown phenocopies miR-200s in inhibiting migration and promoting metastatic colonization. (a) Transwell migration assays were performed in triplicate and is presented as the mean migration of experimental lines as a ratio relative to parental 4TO7 cells ± s.e.m. * P < 0.05 (Student’s t-test). (b) Bar graph showing fold change in number of pulmonary nodules relative to 4TO7 parental line. Data presented as mean ± s.e.m. * P < 0.05 (Student’s t-test). (c) Representative gross lung images and H&E stained lung sections (lower panel) from mice intravenously injected with various cell lines as indicated. Red arrows were used to highlight metastatic nodules, except in Sec23a-KD samples which contained large numbers of nodules. (d) Box plot showing relative expression of SEC23A in ten human primary tumors versus ten metastases. GAPDH was used to normalize expression. P value computed by Student’s t-test. (e) Bar graph showing relative expression of SEC23A in matched primary and metastasis samples collected from six individuals. GAPDH was used to normalize expression. * P < 0.05 (Student’s t-test).
Figure 6
Sec23a knockdown disrupts secretion of proteins that are correlated with suppression of clinical metastasis. (a) Scatter plot showing correlation of secretome profiles between two different Sec23a KD lines and between Sec23a KD and C1+C2 lines. Proteins in common between different lines were used to generate the plots. Orange dots represent proteins reduced in abundance in both conditions; green dots represent proteins increased in abundance in both conditions whereas gray dots represent those proteins that show discordant expression patterns. (b) Kaplan-Meier curves showing relapse-free survival of patients with high and low median expression level of 35 genes reduced in secretion in Sec23a KD lines. (c) Bar graph showing fold increase in number of pulmonary metastases in 4TO7-derived lines stably knocking down Axl, Tinagl1 or Igfbp4 relative to vector control. (d) Representative gross lung images from animals injected via lateral tail vein with various knockdown lines from (c) along with vector control. ** P < 0.01 (Student’s t-test). (e) Kaplan-Meier plots of distant metastasis-free survival of patients in the EMC286 dataset stratified by expression of TINAGL1 (top) or IGFBP4 (bottom). P values were computed by log-rank test. (f) Schematic model of miR-200 function during metastasis. MiR-200s simultaneously target several genes including Zeb1, ZEB2 and Sec23a to inhibit local invasion but promote metastatic colonization. Targeting of Zeb1/2 influences cell-intrinsic epithelial traits whereas targeting of Sec23a modulates tumor-derived secretion of factors such as Igfbp4 and Tinagl1, which influence metastatic colonization by altering tumor-stromal interactions.
Comment in
- The social aspects of EMT-MET plasticity.
Thompson EW, Haviv I. Thompson EW, et al. Nat Med. 2011 Sep 7;17(9):1048-9. doi: 10.1038/nm.2437. Nat Med. 2011. PMID: 21900919 No abstract available.
Similar articles
- Sec23a mediates miR-200c augmented oligometastatic to polymetastatic progression.
Sun Z, Zhou S, Tang J, Ye T, Li J, Liu D, Zhou J, Wang J, Rosie Xing H. Sun Z, et al. EBioMedicine. 2018 Nov;37:47-55. doi: 10.1016/j.ebiom.2018.10.002. Epub 2018 Oct 6. EBioMedicine. 2018. PMID: 30301603 Free PMC article. - MiR-200 Regulates Epithelial-Mesenchymal Transition in Anaplastic Thyroid Cancer via EGF/EGFR Signaling.
Xue L, Su D, Li D, Gao W, Yuan R, Pang W. Xue L, et al. Cell Biochem Biophys. 2015 May;72(1):185-90. doi: 10.1007/s12013-014-0435-1. Cell Biochem Biophys. 2015. PMID: 25542369 Retracted. - miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis.
Ma L, Young J, Prabhala H, Pan E, Mestdagh P, Muth D, Teruya-Feldstein J, Reinhardt F, Onder TT, Valastyan S, Westermann F, Speleman F, Vandesompele J, Weinberg RA. Ma L, et al. Nat Cell Biol. 2010 Mar;12(3):247-56. doi: 10.1038/ncb2024. Epub 2010 Feb 21. Nat Cell Biol. 2010. PMID: 20173740 Free PMC article. - A family of pleiotropically acting microRNAs in cancer progression, miR-200: potential cancer therapeutic targets.
Zhang HF, Xu LY, Li EM. Zhang HF, et al. Curr Pharm Des. 2014;20(11):1896-903. doi: 10.2174/13816128113199990519. Curr Pharm Des. 2014. PMID: 23888967 Review. - The Functions of MicroRNA-200 Family in Ovarian Cancer: Beyond Epithelial-Mesenchymal Transition.
Choi PW, Ng SW. Choi PW, et al. Int J Mol Sci. 2017 Jun 6;18(6):1207. doi: 10.3390/ijms18061207. Int J Mol Sci. 2017. PMID: 28587302 Free PMC article. Review.
Cited by
- A Circle RNA Regulatory Axis Promotes Lung Squamous Metastasis via CDR1-Mediated Regulation of Golgi Trafficking.
Harrison EB, Porrello A, Bowman BM, Belanger AR, Yacovone G, Azam SH, Windham IA, Ghosh SK, Wang M, Mckenzie N, Waugh TA, Van Swearingen AED, Cohen SM, Allen DG, Goodwin TJ, Mascenik T, Bear JE, Cohen S, Randell SH, Massion PP, Major MB, Huang L, Pecot CV. Harrison EB, et al. Cancer Res. 2020 Nov 15;80(22):4972-4985. doi: 10.1158/0008-5472.CAN-20-1162. Epub 2020 Sep 25. Cancer Res. 2020. PMID: 32978168 Free PMC article. - Attenuation of TGF-β signaling suppresses premature senescence in a p21-dependent manner and promotes oncogenic Ras-mediated metastatic transformation in human mammary epithelial cells.
Lin S, Yang J, Elkahloun AG, Bandyopadhyay A, Wang L, Cornell JE, Yeh IT, Agyin J, Tomlinson G, Sun LZ. Lin S, et al. Mol Biol Cell. 2012 Apr;23(8):1569-81. doi: 10.1091/mbc.E11-10-0849. Epub 2012 Feb 22. Mol Biol Cell. 2012. PMID: 22357622 Free PMC article. - Loss of the polarity protein PAR3 activates STAT3 signaling via an atypical protein kinase C (aPKC)/NF-κB/interleukin-6 (IL-6) axis in mouse mammary cells.
Guyer RA, Macara IG. Guyer RA, et al. J Biol Chem. 2015 Mar 27;290(13):8457-68. doi: 10.1074/jbc.M114.621011. Epub 2015 Feb 5. J Biol Chem. 2015. PMID: 25657002 Free PMC article. - Breast Cancer Metastasis.
Kim MY. Kim MY. Adv Exp Med Biol. 2021;1187:183-204. doi: 10.1007/978-981-32-9620-6_9. Adv Exp Med Biol. 2021. PMID: 33983579 - MiR-128 suppresses metastatic capacity by targeting metadherin in breast cancer cells.
Cao D, Zhu H, Zhao Q, Huang J, Zhou C, He J, Liang Y. Cao D, et al. Biol Res. 2020 Sep 29;53(1):43. doi: 10.1186/s40659-020-00311-5. Biol Res. 2020. PMID: 32993809 Free PMC article.
References
- Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. - PubMed
- Thompson EW, Williams ED. EMT and MET in carcinoma--clinical observations, regulatory pathways and new models. Clin Exp Metastasis. 2008;25:591–592. - PubMed
- Yang J, Weinberg RA. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev Cell. 2008;14:818–829. - PubMed
- Chaffer CL, Thompson EW, Williams ED. Mesenchymal to epithelial transition in development and disease. Cells Tissues Organs. 2007;185:7–19. - PubMed
- Jeschke U, Mylonas I, Kuhn C, Shabani N, Kunert-Keil C, Schindlbeck C, Gerber B, Friese K. Expression of E-cadherin in human ductal breast cancer carcinoma in situ, invasive carcinomas, their lymph node metastases, their distant metastases, carcinomas with recurrence and in recurrence. Anticancer Res. 2007;27:1969–1974. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30 CA072720/CA/NCI NIH HHS/United States
- R01 CA141062-03/CA/NCI NIH HHS/United States
- R01 CA141062/CA/NCI NIH HHS/United States
- 090532/Wellcome Trust/United Kingdom
- R01 CA141062-01A1/CA/NCI NIH HHS/United States
- R01 CA134519/CA/NCI NIH HHS/United States
- R01 CA141062-02/CA/NCI NIH HHS/United States
- CRUK_/Cancer Research UK/United Kingdom
- R01 CA134519-04/CA/NCI NIH HHS/United States
- 1R01-CA141062/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous