Filifactor alocis has virulence attributes that can enhance its persistence under oxidative stress conditions and mediate invasion of epithelial cells by porphyromonas gingivalis - PubMed (original) (raw)
Filifactor alocis has virulence attributes that can enhance its persistence under oxidative stress conditions and mediate invasion of epithelial cells by porphyromonas gingivalis
A Wilson Aruni et al. Infect Immun. 2011 Oct.
Abstract
Filifactor alocis, a Gram-positive anaerobic rod, is one of the most abundant bacteria identified in the periodontal pockets of periodontitis patients. There is a gap in our understanding of its pathogenicity and ability to interact with other periodontal pathogens. To evaluate the virulence potential of F. alocis and its ability to interact with Porphyromonas gingivalis W83, several clinical isolates of F. alocis were characterized. F. alocis showed nongingipain protease and sialidase activities. In silico analysis revealed the molecular relatedness of several virulence factors from F. alocis and P. gingivalis. In contrast to P. gingivalis, F. alocis was relatively resistant to oxidative stress and its growth was stimulated under those conditions. Biofilm formation was significantly increased in coculture. There was an increase in adherence and invasion of epithelial cells in coculture compared with P. gingivalis or F. alocis monocultures. In those epithelial cells, endocytic vesicle-mediated internalization was observed only during coculture. The F. alocis clinical isolate had an increased invasive capacity in coculture with P. gingivalis compared to the ATCC 35896 strain. In addition, there was variation in the proteomes of the clinical isolates compared to the ATCC 35896 strain. Hypothetical proteins and those known to be important virulence factors in other bacteria were identified. These results indicate that F. alocis has virulence properties that may enhance its ability to survive and persist in the periodontal pocket and may play an important role in infection-induced periodontal disease.
Figures
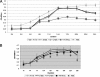
Fig. 1.
(A) Growth rates of Filifactor alocis strains. Cultures of F. alocis and P. gingivalis were incubated at 37°C under anaerobic conditions. Growth rates were determined by measuring OD600 over time. The data are means ± standard deviations (SD) of three independent experiments. The generation time (G) was calculated using the equation G = t/n, where t is the time interval and n is the number of generations. n = (log Nt − log _N_0)/log 2, where _N_0 is the number of bacteria at culture start time and Nt is the number of bacterial at the end of log phase. FA-ATCC, F. alocis ATCC 35896. (B) Growth of Filifactor alocis under oxidative stress. F. alocis and P. gingivalis W83 strains grown to early log phase were treated with 0.25 mM H2O2 and further incubated for 24 h. The results shown are representative of three independent replicates.
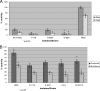
Fig. 2.
Comparison of gingipain, nongingipain protease, and sialidase activities of F. alocis strains and P. gingivalis W83. F. alocis and P. gingivalis were grown to exponential phase (OD600 of 0.8). (A) Gingipain activities of F. alocis and P. gingivalis strains were tested against BAPNA (_N_-α-benzoyl-
l
-tyrosine _p_-nitroanilide; Rgp) and ALNA (acetyl-Lys-_p_-nitroanilide-HCl; Kgp) using whole-cell culture. The results shown are from three independent experiments. The clinical strains are statistically compared with the ATCC strain. (B) Nongingipain protease and sialidase activities of F. alocis strains. Nongingipain protease activity was estimated using a FRET-based assay method. Sialidase activity was tested using the Amplex Red neuraminidase assay kit. Percent activity was determined using the standard supplied with the kit. The results shown are from three independent experiments. Filifactor alocis strains are statistically compared with P. gingivalis.
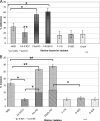
Fig. 3.
Amino acid utilization by Filifactor alocis strains. Amino acids arginine, lysine, valine, threonine, isoleucine, glycine, and cystine were used at 100 μM concentration in BHI medium supplemented with hemin (5 μg/ml), vitamin K (0.5 μg/ml), and cysteine (0.1%). The cultures were grown for 8 h, and OD600 was determined. F. alocis grown without any amino acid supplementation was used as a control. The data are means ± SD of three independent experiments. Arginine supplementation was compared statistically with the other amino acids used.
Fig. 4.
Biofilm formation by mono- and cocultures of Filifactor alocis and P. gingivalis ATCC 33277 (PG). Overnight cultures of F. alocis and/or P. gingivalis were diluted with fresh BHI medium to obtain a concentration of 5 × 107 CFU/ml. The cells were aliquoted to a 96-well microtiter plate (260 μl per well) and incubated anaerobically at 37°C for 24 h. The biofilms were stained with crystal violet. The solubilized crystal violet was transferred to a new microtiter plate, and the OD540 was determined spectrophotometrically. Medium and P. gingivalis W83 were used as controls. Coculture was statistically compared with P. gingivalis monoculture.
Fig. 5.
(A) Adherence by mono- and cocultures of Filifactor alocis strains and P. gingivalis W83 in epithelial cells. HeLa cells were grown and maintained at 37°C under 5% CO2 in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum with antibiotics. Confluent stock cultures were trypsinized, adjusted to approximately 5 × 103 cells/ml, seeded (1 ml per well) into 12-well plates, and further incubated for 48 h to reach semiconfluence. Epithelial cells (105 cells) were infected with 107 bacteria (MOI = 100). Error bars represent mean ± SD of four independent experiments. Statistical significance was determined using analysis of variance (ANOVA) with the Bonferroni posttest. FA-ATCC, F. alocis ATCC 35896; PG, P. gingivalis; FAC, F. alocis clinical strain D-62D. (B) Invasion by mono- and cocultures of Filifactor alocis strains and P. gingivalis W83 into epithelial cells. Bacteria were grown to exponential phase in BHI broth, washed three times in PBS, and adjusted to 107 CFU/ml in DMEM. The epithelial cell monolayer was washed and infected with bacteria at an MOI of 1:100 (105 epithelial cells) and then incubated at 37°C for 45 min under 5% CO2. Error bars represent mean ± SD of four independent experiments. Statistical significance was determined using ANOVA with the Bonferroni posttest.
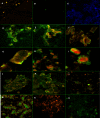
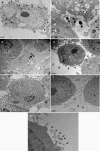
Fig. 6.
Fluorescence in situ hybridization. FISH was performed using a trypsinized culture of HeLa cells (5 × 103 cells/ml) grown on coverslips. Probe-hybridized slides were air dried, mounted using the SlowFade Gold antifade reagent, and visualized using an inverted Axio Observer Z1 confocal microscope. The images were analyzed using Zeiss LSM 710 software. The images shown are representative of more than 12 viewing fields under a given magnification. F. alocis D-62D and P. gingivalis W83 were used in this study. (A) P. gingivalis W83 control cells showing specific red-orange fluorescence (645-nm absorbance/665-nm emission) (magnification, ×20). (B) F. alocis D-62D control cells showing specific green fluorescence (549-nm absorbance/563-nm emission) (magnification, ×20). (C) Negative control showing no fluorescence using DAPI (magnifi- cation, ×20). (D) Coinfection producing marginal green fluorescence around the epithelial cells, showing adherence of F. alocis D-62D seen after 20 min p.i. (magnification, ×40). (E) Coinfection producing a shift in the fluorescence, showing invasion by P. gingivalis W83 over F. alocis D-62D at 30 min p.i. (magnification, ×40). (F) Coinfected epithelial cells showing peripheral adherence of F. alocis D-62D and early invasion by P. gingivalis W83 (magnification, ×40) at 45 min p.i. (G) Coinfected individual epithelial cell showing early invasion by P. gingivalis W83 (magnification, ×63) at 45 min p.i. (H) Coinfected individual epithelial cell showing early invasion by P. gingivalis W83 (intense red-orange fluorescence in the cytoplasm) (magnification, ×40) at 45 min p.i. (I) Coinfected epithelial cell showing sparse adherence of F. alocis D-62D and cytoplasm showing the invading P. gingivalis W83 (magnification, ×40) at 45 min p.i. (J) Coinfection of F. alocis D-62D and P. gingivalis W83 showing generalized invasion of epithelial cells at 1 h p.i. (magnification, ×20). (K) Coinfection of F. alocis D-62D and P. gingivalis W83 showing general invasion of epithelial cells at 1 h p.i. (magnification, ×40). (L and M) Equal invasion was noticed after 1 h p.i. (magnification, ×20). (N) Invasion by P. gingivalis W83 monoculture in epithelial cells (magnification, ×20). (O) Invasion by F. alocis monoculture in epithelial cells (magnification, ×20).
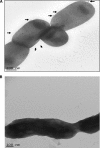
Fig. 7.
Transmission electron microscopy was performed using the FEI Tecnai G2 TEM and Formvar carbon-coated grids. Conventional electron microscopy was performed by staining the sample with 1% phosphotungstic acid. (A). The external surface of Filifactor alocis ATCC 35896 shows rudimentary markings and slender projections resembling pili (arrows). (B) Filifactor alocis clinical strain D-62D showing thick slimy outer cover with no external projections or rudimentary marks.
Fig. 8.
Ultrathin sections of the infected epithelial cell monolayer were washed with PBS, fixed with 2.5% glutaraldehyde, and stained with lead citrate and uranyl acetate. The grids were examined using by the FEI Tecnai G2 transmission electron microscope. Arrows show vesicles in the cytoplasm containing both Filifactor alocis D-62D and P. gingivalis W83. (A) Ultrathin sections of the coculture-infected epithelial cell monolayer at 45 min p.i. Arrows show endocytic vesicles in the cytoplasm containing both bacteria. (B) Monoculture of P. gingivalis W83 at 45 min p.i., showing invading organisms without an external envelope or covering. (C) Monoculture of Filifactor alocis D-62D at 45 min p.i., showing invading organisms without a covering. (D) Invasion by a coculture of F. alocis D-62D and P. gingivalis W83 at 30 min p.i., showing formation of the vesicular covering (arrows). The covering was noticed when the bacteria were in contact with the epithelial cells. Such receptor-mediated endocytosis was found only during coculture. (E) Coculture of P. gingivalis W83 and F. alocis D-62D at 30 min p.i., showing a higher number of invading P. gingivalis W83 cells. (F and G) P. gingivalis W83 monoculture at 30 min p.i., showing cytoskeleton variation through formation of microvilli, filamentous projections of the epithelial cells facilitating invasion by the bacteria.
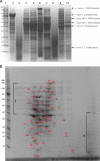
Fig. 9.
(A) SDS-PAGE of cell fractions of F. alocis. Cell fractions of F. alocis strains were analyzed by SDS-PAGE and stained using Coomassie SimplyBlue safe stain. Approximately 25 μg of protein was loaded onto each lane. Lane 1, molecular mass (kDa) markers; lane 2, particle-free pellet (ATCC 35896); lane 3, particle-free supernatant (ATCC 35896); lane 4, total cell lysate (ATCC 35896); lane 5, cytosolic fraction (ATCC 35896); lane 6, membrane fraction (ATCC 35896); lane 7, total cell lysate (D-62D); lane 8, particle-free supernatant (D-62D); lane 9, cytosolic fraction (D-62D); lane 10, membrane fraction (D-62D). (B) 2D gel electrophoresis of total cell lysate of F. alocis (ATCC 35896). 2D gel electrophoresis was carried out using 7-cm gel strips in a Protean IEF cell. The protein sample was diluted to a final concentration of 350 μg protein, and 20 μl of the sample was added and was run in two dimensions. After equilibration, the IPG strips were loaded onto the gel, electrophoresed at 200 V and 0.3 A for 4 to 5 h, and stained with Coomassie SimplyBlue safe strain. §, poorly resolved spots in the range of 30 to 80 kDa; ¥, poorly resolved spots in the range of 10 to 40 kDa. Numbers correspond to the discrete protein spots (see Table S1 in the supplemental material).
Similar articles
- Role of the Filifactor alocis Hypothetical Protein FA519 in Oxidative Stress Resistance.
Aja E, Mishra A, Dou Y, Fletcher HM. Aja E, et al. Microbiol Spectr. 2021 Dec 22;9(3):e0121221. doi: 10.1128/Spectrum.01212-21. Epub 2021 Nov 10. Microbiol Spectr. 2021. PMID: 34756068 Free PMC article. - Proteome analysis of coinfection of epithelial cells with Filifactor alocis and Porphyromonas gingivalis shows modulation of pathogen and host regulatory pathways.
Aruni AW, Zhang K, Dou Y, Fletcher H. Aruni AW, et al. Infect Immun. 2014 Aug;82(8):3261-74. doi: 10.1128/IAI.01727-14. Epub 2014 May 27. Infect Immun. 2014. PMID: 24866790 Free PMC article. - Proteome variation among Filifactor alocis strains.
Aruni AW, Roy F, Sandberg L, Fletcher HM. Aruni AW, et al. Proteomics. 2012 Nov;12(22):3343-64. doi: 10.1002/pmic.201200211. Proteomics. 2012. PMID: 23008013 Free PMC article. - Filifactor alocis: The Newly Discovered Kid on the Block with Special Talents.
Aruni W, Chioma O, Fletcher HM. Aruni W, et al. J Dent Res. 2014 Aug;93(8):725-32. doi: 10.1177/0022034514538283. Epub 2014 Jun 4. J Dent Res. 2014. PMID: 24898946 Free PMC article. Review.
Cited by
- Filifactor alocis enhances survival of Porphyromonas gingivalis W83 in response to H2 O2 -induced stress.
Mishra A, Dou Y, Wang C, Fletcher HM. Mishra A, et al. Mol Oral Microbiol. 2024 Feb;39(1):12-26. doi: 10.1111/omi.12445. Epub 2023 Dec 1. Mol Oral Microbiol. 2024. PMID: 38041478 Free PMC article. - Whole Genome Sequencing and Phenotypic Analysis of Antibiotic Resistance in Filifactor alocis Isolates.
Romero-Martínez R, Maher A, Àlvarez G, Figueiredo R, León R, Arredondo A. Romero-Martínez R, et al. Antibiotics (Basel). 2023 Jun 15;12(6):1059. doi: 10.3390/antibiotics12061059. Antibiotics (Basel). 2023. PMID: 37370380 Free PMC article. - Filifactor alocis and Tumor Necrosis Factor-Alpha Stimulate Synthesis of Visfatin by Human Macrophages.
Nogueira AVB, Nokhbehsaim M, Damanaki A, Eick S, Kirschneck C, Schröder A, Jantsch J, Deschner J. Nogueira AVB, et al. Int J Mol Sci. 2021 Jan 27;22(3):1235. doi: 10.3390/ijms22031235. Int J Mol Sci. 2021. PMID: 33513808 Free PMC article. - Porphyromonas gingivalis: where do we stand in our battle against this oral pathogen?
Howard KC, Gonzalez OA, Garneau-Tsodikova S. Howard KC, et al. RSC Med Chem. 2021 Feb 26;12(5):666-704. doi: 10.1039/d0md00424c. eCollection 2021 May 26. RSC Med Chem. 2021. PMID: 34124669 Free PMC article. Review. - Microbiota in Human Periodontal Abscess Revealed by 16S rDNA Sequencing.
Chen J, Wu X, Zhu D, Xu M, Yu Y, Yu L, Zhang W. Chen J, et al. Front Microbiol. 2019 Jul 30;10:1723. doi: 10.3389/fmicb.2019.01723. eCollection 2019. Front Microbiol. 2019. PMID: 31417518 Free PMC article.
References
- Amano A. 2007. Disruption of epithelial barrier and impairment of cellular function by Porphyromonas gingivalis. Front. Biosci. 12:3965–3974 - PubMed
- Amano A., Furuta N., Tsuda K. 2010. Host membrane trafficking for conveyance of intracellular oral pathogens. Periodontol. 2000 52:84–93 - PubMed
- Byers H. L., Homer K. A., Tarelli E., Beighton D. 1999. N-acetylneuraminic acid transport by Streptococcus oralis strain AR3. J. Med. Microbiol. 48:375–381 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 DE019730/DE/NIDCR NIH HHS/United States
- DE-13664/DE/NIDCR NIH HHS/United States
- R01 DE013664/DE/NIDCR NIH HHS/United States
- R56 DE013664/DE/NIDCR NIH HHS/United States
- R21 DE022724/DE/NIDCR NIH HHS/United States
- DE-019730/DE/NIDCR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous