Expansion of a unique CD57⁺NKG2Chi natural killer cell subset during acute human cytomegalovirus infection - PubMed (original) (raw)
. 2011 Sep 6;108(36):14725-32.
doi: 10.1073/pnas.1110900108. Epub 2011 Aug 8.
Jeffrey M Milush, Brian S Schwartz, Marcelo J Pando, Jessica Jarjoura, Vanessa A York, Jeffrey P Houchins, Steve Miller, Sang-Mo Kang, Phillip J Norris, Douglas F Nixon, Lewis L Lanier
Affiliations
- PMID: 21825173
- PMCID: PMC3169160
- DOI: 10.1073/pnas.1110900108
Expansion of a unique CD57⁺NKG2Chi natural killer cell subset during acute human cytomegalovirus infection
Sandra Lopez-Vergès et al. Proc Natl Acad Sci U S A. 2011.
Abstract
During human CMV infection, there is a preferential expansion of natural killer (NK) cells expressing the activating CD94-NKG2C receptor complex, implicating this receptor in the recognition of CMV-infected cells. We hypothesized that NK cells expanded in response to pathogens will be marked by expression of CD57, a carbohydrate antigen expressed on highly mature cells within the CD56(dim)CD16(+) NK cell compartment. Here we demonstrate the preferential expansion of a unique subset of NK cells coexpressing the activating CD94-NKG2C receptor and CD57 in CMV(+) donors. These CD57(+)NKG2C(hi) NK cells degranulated in response to stimulation through their NKG2C receptor. Furthermore, CD57(+)NKG2C(hi) NK cells preferentially lack expression of the inhibitory NKG2A receptor and the inhibitory KIR3DL1 receptor in individuals expressing its HLA-Bw4 ligand. Moreover, in solid-organ transplant recipients with active CMV infection, the percentage of CD57(+)NKG2C(hi) NK cells in the total NK cell population preferentially increased. During acute CMV infection, the NKG2C(+) NK cells proliferated, became NKG2C(hi), and finally acquired CD57. Thus, we propose that CD57 might provide a marker of "memory" NK cells that have been expanded in response to infection.
Conflict of interest statement
Conflict of interest statement: J.P.H. is an employee of R&D Systems. The other authors have no conflicts.
Figures
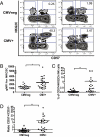
Fig. 1.
A unique population of CD57+NKG2Chi NK cells is detected in CMV+ donors. (A) CD3negCD56dimCD16+ NK cells from four CMV− (Upper) and CMV+ (Lower) donors were gated and analyzed for expression of CD57 and NKG2C. (B) The geometric mean fluorescence intensity (gMFI) for NKG2C in the CD57+ NK cell subset from CMV− and CMV+ donors is shown. (C) The percentage of CD57+NKG2Chi cells in the CD56dimCD16+ NK cell subset from CMV+ and CMV− donors is shown. (D) The ratio of CD57+NKG2Chi/CD57−NKG2Chi NK cells for each CMV− and CMV+ donors is shown. Triangles represent CMV− donors (n = 23), circles represent CMV+ donors (i.e., seropositive and/or CMV-specific T-cell responses; n = 29).
Fig. 2.
CD57+NKG2Chi NK cells from CMV+ donors degranulated when activated through NKG2C. PBMCs from CMV+ donors where stimulated with plate-bound antibodies against CD56 (negative control), NKG2C, or CD16 (positive control). The percentage of CD107α+ cells was determined for CD57+NKG2Chi NK cells (n = 7).
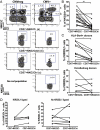
Fig. 3.
Certain inhibitory NK receptors are underrepresented on the CD57+NKG2Chi NK cell subset from CMV+ donors. (A) CD56dimCD16+ NK cells from CMV− (Left) and CMV+ (Right) donors were gated and analyzed for expression of CD57 and NKG2C. CD57+NKG2C− (a), CD57+NKG2Cdim (b), and CD57+NKG2Chi (c) NK cells were assessed for CD56 and NKG2A expression. (B) The percentage of NKG2A+ cells was determined on CD57+NKG2C− and CD57+NKG2Chi NK cells from CMV+ donors (n = 16). (C) The percentage of KIR3DL1+ cells was determined on CD57+NKG2C− and CD57+NKG2Chi NK cells from CMV+ donors. Graphs from HLA-Bw4+ (Upper; n = 9) and HLA-Bw4− donors (Lower; n = 6) are shown. (D) The percentage of KIR2DL1+ cells was determined on CD57+NKG2C− and CD57+NKG2Chi NK cells for CMV+ donors who have the KIR2DL1 ligand (Left; n = 5) or do not (Right; n = 7).
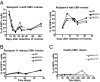
Fig. 4.
CD57+NKG2Chi NK cells increase during acute CMV infection in SOT recipients. (A) NKG2C and CD57 expression on CD56dimCD16+ NK cells from SOT recipients with acute CMV infection was analyzed. Antiviral treatment was administrated immediately after detection of viremia (day 0). The percentage of CD57+NKG2Chi NK cells detected over time and the CMV viral load detected in plasma (value > 400 copies/mL) from representative patients are shown (n = 6). Solid line and square represent CD57+NKG2Chi NK cells, dashed line and triangle represent CMV DNA. (B and C) The percentages of CD57+NKG2Chi NK cells detected over time from one representative SOT recipient without CMV viremia (B; n = 5) and from a healthy CMV+ donor (C) are shown. (D and E) Approximate absolute numbers (in 103 cells/L) of CD57+NKG2Chi NK cells detected over time in one representative SOT recipient with CMV viremia (D; CMV viral load shown; n = 6) and in SOT recipients without CMV viremia (E; n = 5).
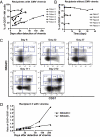
Fig. 5.
During CMV acute infection, NKG2C+ NK cells acquire CD57 expression and become CD57+NKG2Chi NK cells. (A and B) The percentages of CD57+ NK cells detected over time in SOT recipients with acute CMV infection (A; n = 6) or without CMV viremia (B; n = 5) are shown. (C) CD56dimCD16+ NK cells were analyzed for expression of CD57 and NKG2C. Plots for different time points from a representative SOT recipient with CMV viremia are shown (n = 6). Population gates: CD57−NKG2Cdim, CD57−NKG2Chi, CD57dimNKG2Chi, and CD57+NKG2Chi NK cells. (D) CD56dimCD16+ NK cells were gated on NKG2C+ and NKG2C− cells and the percentage of CD57+ cells for each subset was assessed over time. The ratio of CD57+/CD57− cells on each subset from a representative SOT recipient with CMV viremia is shown (n = 6). Solid square is from NKG2C+, solid triangle is from NKG2C− NK cells.
Fig. 6.
All NK cells divide in response to acute CMV infection. (A and B) CD56dimCD16+ NK cells were gated on NKG2C+ and NKG2C− cells, and Ki-67 expression on all CD56dimCD16+ NK cells and in each subset was assessed. Graphs from representative SOT recipients with CMV viremia (A; n = 4) and without CMV viremia (B) are shown. Solid line and empty circle represent CD56dimCD16+ NK cells, solid line and square represent NKG2C+ NK cells, and dashed line and triangle represent NKG2C− NK cells. (C) The percentage of Ki-67+ cells within the CD56dimCD16+ NK cells detected over time in healthy CMV+ donors is shown.
Similar articles
- Cutting edge: NKG2C(hi)CD57+ NK cells respond specifically to acute infection with cytomegalovirus and not Epstein-Barr virus.
Hendricks DW, Balfour HH Jr, Dunmire SK, Schmeling DO, Hogquist KA, Lanier LL. Hendricks DW, et al. J Immunol. 2014 May 15;192(10):4492-6. doi: 10.4049/jimmunol.1303211. Epub 2014 Apr 16. J Immunol. 2014. PMID: 24740502 Free PMC article. Clinical Trial. - Cytomegalovirus reactivation after allogeneic transplantation promotes a lasting increase in educated NKG2C+ natural killer cells with potent function.
Foley B, Cooley S, Verneris MR, Pitt M, Curtsinger J, Luo X, Lopez-Vergès S, Lanier LL, Weisdorf D, Miller JS. Foley B, et al. Blood. 2012 Mar 15;119(11):2665-74. doi: 10.1182/blood-2011-10-386995. Epub 2011 Dec 16. Blood. 2012. PMID: 22180440 Free PMC article. - Human cytomegalovirus (CMV)-induced memory-like NKG2C(+) NK cells are transplantable and expand in vivo in response to recipient CMV antigen.
Foley B, Cooley S, Verneris MR, Curtsinger J, Luo X, Waller EK, Anasetti C, Weisdorf D, Miller JS. Foley B, et al. J Immunol. 2012 Nov 15;189(10):5082-8. doi: 10.4049/jimmunol.1201964. Epub 2012 Oct 17. J Immunol. 2012. PMID: 23077239 Free PMC article. Clinical Trial. - Adaptive reconfiguration of the human NK-cell compartment in response to cytomegalovirus: a different perspective of the host-pathogen interaction.
Muntasell A, Vilches C, Angulo A, López-Botet M. Muntasell A, et al. Eur J Immunol. 2013 May;43(5):1133-41. doi: 10.1002/eji.201243117. Eur J Immunol. 2013. PMID: 23552990 Review. - The CD94/NKG2C+ NK-cell subset on the edge of innate and adaptive immunity to human cytomegalovirus infection.
López-Botet M, Muntasell A, Vilches C. López-Botet M, et al. Semin Immunol. 2014 Apr;26(2):145-51. doi: 10.1016/j.smim.2014.03.002. Epub 2014 Mar 22. Semin Immunol. 2014. PMID: 24666761 Review.
Cited by
- Mucosal-homing natural killer cells are associated with aging in persons living with HIV.
Kroll KW, Shah SV, Lucar OA, Premeaux TA, Shikuma CM, Corley MJ, Mosher M, Woolley G, Bowler S, Ndhlovu LC, Reeves RK. Kroll KW, et al. Cell Rep Med. 2022 Oct 18;3(10):100773. doi: 10.1016/j.xcrm.2022.100773. Epub 2022 Oct 7. Cell Rep Med. 2022. PMID: 36208628 Free PMC article. - NK Cell Memory to Cytomegalovirus: Implications for Vaccine Development.
Forrest C, Gomes A, Reeves M, Male V. Forrest C, et al. Vaccines (Basel). 2020 Jul 20;8(3):394. doi: 10.3390/vaccines8030394. Vaccines (Basel). 2020. PMID: 32698362 Free PMC article. Review. - Natural killer cell therapies.
Vivier E, Rebuffet L, Narni-Mancinelli E, Cornen S, Igarashi RY, Fantin VR. Vivier E, et al. Nature. 2024 Feb;626(8000):727-736. doi: 10.1038/s41586-023-06945-1. Epub 2024 Feb 21. Nature. 2024. PMID: 38383621 Review. - Exploiting innate immunity for cancer immunotherapy.
Yi M, Li T, Niu M, Mei Q, Zhao B, Chu Q, Dai Z, Wu K. Yi M, et al. Mol Cancer. 2023 Nov 27;22(1):187. doi: 10.1186/s12943-023-01885-w. Mol Cancer. 2023. PMID: 38008741 Free PMC article. Review. - Multiplex and genome-wide analyses reveal distinctive properties of KIR+ and CD56+ T cells in human blood.
Chan WK, Rujkijyanont P, Neale G, Yang J, Bari R, Das Gupta N, Holladay M, Rooney B, Leung W. Chan WK, et al. J Immunol. 2013 Aug 15;191(4):1625-36. doi: 10.4049/jimmunol.1300111. Epub 2013 Jul 15. J Immunol. 2013. PMID: 23858032 Free PMC article.
References
- Biron CA, Byron KS, Sullivan JL. Severe herpesvirus infections in an adolescent without natural killer cells. N Engl J Med. 1989;320:1731–1735. - PubMed
- Orange JS. Human natural killer cell deficiencies and susceptibility to infection. Microbes Infect. 2002;4:1545–1558. - PubMed
- Etzioni A, et al. Fatal varicella associated with selective natural killer cell deficiency. J Pediatr. 2005;146:423–425. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- AI068129/AI/NIAID NIH HHS/United States
- HL095470/HL/NHLBI NIH HHS/United States
- R01 HL095470/HL/NHLBI NIH HHS/United States
- P30 AI027763/AI/NIAID NIH HHS/United States
- R01 AI068129/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous