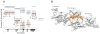Oligomeric structure of the chemokine CCL5/RANTES from NMR, MS, and SAXS data - PubMed (original) (raw)
Oligomeric structure of the chemokine CCL5/RANTES from NMR, MS, and SAXS data
Xu Wang et al. Structure. 2011.
Abstract
CCL5 (RANTES) is a proinflammatory chemokine known to activate leukocytes through its receptor, CCR5. Although the monomeric form of CCL5 is sufficient to cause cell migration in vitro, CCL5's propensity for aggregation is essential for migration in vivo, T cell activation and apoptosis, and HIV entry into cells. However, there is currently no structural information on CCL5 oligomers larger than the canonical CC chemokine dimer. In this study the solution structure of a CCL5 oligomer was investigated using an integrated approach, including NMR residual dipolar couplings to determine allowed relative orientations of the component monomers, SAXS to restrict overall shape, and hydroxyl radical footprinting and NMR cross-saturation experiments to identify interface residues. The resulting model of the CCL5 oligomer provides a basis for explaining the disaggregating effect of E66 and E26 mutations and suggests mechanisms by which glycosaminoglycan binding may promote oligomer formation and facilitate cell migration in vivo.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures
Figure 1
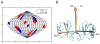
A) Sauson-Flamsteed projection plot of alignment frame orientations for WT CCL5 aligned in both positively charged and neutral polyacrylamide gel. The X-axes of the two frames share a common orientation. B) Orientation of the alignment tensor relative to the CCL5 dimer. The X-axis of the principal axes of the alignment tensor is shown in red, the Y-axis is shown in blue and the Z-axis is in orange. The golden arrow indicates the orientation of the symmetry axis from the dimer crystal structure. (see also Figure S2 and S3)
Figure 2
A) Fitting to SAXS scattering data for model from grid point 19×13, χ=1.13, 40% of the protein is assumed in be in the tetramer form. B) Contour map of the SAXS fitting χ values of the models generated on the grid. Colors ranging from blue to yellow represent high to low χ values. (see also Figure S4)
Figure 3
A) Contour maps of the combined residue pair and SAXS scores for each model on the grid. B) Model from grid point 19×13, which is representative of the models from the group with the best (orange region) combined score. (see also Figure S5)
Figure 4
A) Plot of residue specific cross saturation-induced amide proton signal intensity changes for WT and E66S CCL5. B) Surface plot of the CCL5 tetramer model with residues identified as being specifically perturbed in the wild type (residues 26 to 29, 33, 34, 66 and 67) colored in orange and the dimer interface (residues 6 to 10) colored in red.
Figure 5
A) Plot of residue specific hydroxyl radical modification percentage for WT and E66S CCL5. The degree of modification is analyzed at the peptide level, and the major sites of oxidation are identified at the residue level for each peptide. Residues identified as being protected from modification are indicated. B) Surface plot of the CCL5 tetramer with residues identified as being in the tetrameric interface (residues 26 to 29, 41, 61, 62, 67, 68,) colored orange. (see also Figure S6)
Figure 6
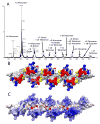
A) Native spray mass spectrum of WT CCL5 (10 μM) at pH 4.5. Even-numbered oligomers from dimer to octamer are observed, indicating that the oligomer is built from a concatenation of dimer substructures. B) Surface plot of the CCL5 octamer model with residues perturbed by GAGs (residues 44–48, 55, 56) shown in red and residues known to contact CCR5 at pH 6 (residues 16, 17, 21, 23) shown in blue. The N-terminus of CCL5, which is both perturbed by GAGs and known to bind to CCR5 in CCL5 monomer is colored yellow. C) Electrostatic potential plot of the octamer surface showing large patches of basic regions (blue) throughout the protein.
Figure 7
A) Details of the inter-dimer hydrophobic interactions in model 19×13. The hydrophobic interface is formed by Y27, F28, I62 and L65. B) Electrostatic interactions at the dimer-dimer interface. K25, E26, E66 & R44 can form pairs of electrostatic bonds. C) Ribbon representations of the proposed CCL5 tetramer (red) and the MIP1α tetramer (blue). A single dimer unit from each tetramer is arranged in identical orientation.
Similar articles
- Integrative Model to Coordinate the Oligomerization and Aggregation Mechanisms of CCL5.
Chen YC, Chen SP, Li JY, Chen PC, Lee YZ, Li KM, Zarivach R, Sun YJ, Sue SC. Chen YC, et al. J Mol Biol. 2020 Feb 14;432(4):1143-1157. doi: 10.1016/j.jmb.2019.12.049. Epub 2020 Jan 11. J Mol Biol. 2020. PMID: 31931012 - Structural basis for oligomerization and glycosaminoglycan binding of CCL5 and CCL3.
Liang WG, Triandafillou CG, Huang TY, Zulueta MM, Banerjee S, Dinner AR, Hung SC, Tang WJ. Liang WG, et al. Proc Natl Acad Sci U S A. 2016 May 3;113(18):5000-5. doi: 10.1073/pnas.1523981113. Epub 2016 Apr 18. Proc Natl Acad Sci U S A. 2016. PMID: 27091995 Free PMC article. - Recognition of RANTES by extracellular parts of the CCR5 receptor.
Duma L, Häussinger D, Rogowski M, Lusso P, Grzesiek S. Duma L, et al. J Mol Biol. 2007 Jan 26;365(4):1063-75. doi: 10.1016/j.jmb.2006.10.040. Epub 2006 Oct 17. J Mol Biol. 2007. PMID: 17101151 - Structural NMR of protein oligomers using hybrid methods.
Wang X, Lee HW, Liu Y, Prestegard JH. Wang X, et al. J Struct Biol. 2011 Mar;173(3):515-29. doi: 10.1016/j.jsb.2010.11.005. Epub 2010 Nov 11. J Struct Biol. 2011. PMID: 21074622 Free PMC article. Review. - Structural characterization of proteins and complexes using small-angle X-ray solution scattering.
Mertens HD, Svergun DI. Mertens HD, et al. J Struct Biol. 2010 Oct;172(1):128-41. doi: 10.1016/j.jsb.2010.06.012. Epub 2010 Jun 15. J Struct Biol. 2010. PMID: 20558299 Review.
Cited by
- Chemokine Receptor N-Terminus Charge Dictates Reliance on Post-Translational Modifications for Effective Ligand Capture and Following Boosting by Defense Peptides.
Xu T, Schou AS, Lackman JJ, Barrio-Calvo M, Verhallen L, Goth CK, Jensen BAH, Veldkamp CT, Volkman BF, Peterson FC, Hjortø GM. Xu T, et al. Int J Mol Sci. 2024 Oct 9;25(19):10854. doi: 10.3390/ijms251910854. Int J Mol Sci. 2024. PMID: 39409188 Free PMC article. - Chemokine Binding to Tenascin-C Influences Chemokine-Induced Immune Cell Migration.
Domaingo A, Jokesch P, Schweiger A, Gschwandtner M, Gerlza T, Koch M, Midwood KS, Kungl AJ. Domaingo A, et al. Int J Mol Sci. 2023 Sep 28;24(19):14694. doi: 10.3390/ijms241914694. Int J Mol Sci. 2023. PMID: 37834140 Free PMC article. - Heterologous Interactions with Galectins and Chemokines and Their Functional Consequences.
Mayo KH. Mayo KH. Int J Mol Sci. 2023 Sep 14;24(18):14083. doi: 10.3390/ijms241814083. Int J Mol Sci. 2023. PMID: 37762385 Free PMC article. Review. - Heterodimers Are an Integral Component of Chemokine Signaling Repertoire.
Kaffashi K, Dréau D, Nesmelova IV. Kaffashi K, et al. Int J Mol Sci. 2023 Jul 19;24(14):11639. doi: 10.3390/ijms241411639. Int J Mol Sci. 2023. PMID: 37511398 Free PMC article. Review. - Chemokine Heteromers and Their Impact on Cellular Function-A Conceptual Framework.
Blanchet X, Weber C, von Hundelshausen P. Blanchet X, et al. Int J Mol Sci. 2023 Jun 30;24(13):10925. doi: 10.3390/ijms241310925. Int J Mol Sci. 2023. PMID: 37446102 Free PMC article. Review.
References
- Al-Hashimi HM, Bolon PJ, Prestegard JH. Molecular symmetry as an aid to geometry determination in ligand protein complexes. J Magn Reson. 2000;142:153–158. - PubMed
- Alon R, Grabovsky V, Feigelson S. Chemokine induction of integrin adhesiveness on rolling and arrested leukocytes local signaling events or global stepwise activation? Microcirculation. 2003;10:297–311. - PubMed
- Appay V, Brown A, Cribbes S, Randle E, Czaplewski LG. Aggregation of RANTES is responsible for its inflammatory properties - Characterization of nonaggregating, noninflammatory RANTES mutants. Journal of Biological Chemistry. 1999;274:27505–27512. - PubMed
- Baltus T, Weber KS, Johnson Z, Proudfoot AE, Weber C. Oligomerization of RANTES is required for CCR1-mediated arrest but not CCR5-mediated transmigration of leukocytes on inflamed endothelium. Blood. 2003;102:1985–1988. - PubMed
- Braunersreuther V, Pellieux C, Pelli G, Burger F, Steffens S, Montessuit C, Weber C, Proudfoot A, Mach F, Arnaud C. Chemokine CCL5/RANTES inhibition reduces myocardial reperfusion injury in atherosclerotic mice. J Mol Cell Cardiol. 2010;48:789–798. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI037113-13S1/AI/NIAID NIH HHS/United States
- P41-RR005351/RR/NCRR NIH HHS/United States
- R01AI37113/AI/NIAID NIH HHS/United States
- R00 GM088483/GM/NIGMS NIH HHS/United States
- P41 RR005351-22/RR/NCRR NIH HHS/United States
- K99GM088483/GM/NIGMS NIH HHS/United States
- K99 GM088483/GM/NIGMS NIH HHS/United States
- R00 GM088483-03/GM/NIGMS NIH HHS/United States
- R01 AI037113-13/AI/NIAID NIH HHS/United States
- P41 GM103390/GM/NIGMS NIH HHS/United States
- R01 AI037113/AI/NIAID NIH HHS/United States
- P41 RR005351/RR/NCRR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases