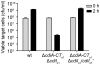Identification of functional toxin/immunity genes linked to contact-dependent growth inhibition (CDI) and rearrangement hotspot (Rhs) systems - PubMed (original) (raw)
Identification of functional toxin/immunity genes linked to contact-dependent growth inhibition (CDI) and rearrangement hotspot (Rhs) systems
Stephen J Poole et al. PLoS Genet. 2011 Aug.
Abstract
Bacterial contact-dependent growth inhibition (CDI) is mediated by the CdiA/CdiB family of two-partner secretion proteins. Each CdiA protein exhibits a distinct growth inhibition activity, which resides in the polymorphic C-terminal region (CdiA-CT). CDI(+) cells also express unique CdiI immunity proteins that specifically block the activity of cognate CdiA-CT, thereby protecting the cell from autoinhibition. Here we show that many CDI systems contain multiple cdiA gene fragments that encode CdiA-CT sequences. These "orphan" cdiA-CT genes are almost always associated with downstream cdiI genes to form cdiA-CT/cdiI modules. Comparative genome analyses suggest that cdiA-CT/cdiI modules are mobile and exchanged between the CDI systems of different bacteria. In many instances, orphan cdiA-CT/cdiI modules are fused to full-length cdiA genes in other bacterial species. Examination of cdiA-CT/cdiI modules from Escherichia coli EC93, E. coli EC869, and Dickeya dadantii 3937 confirmed that these genes encode functional toxin/immunity pairs. Moreover, the orphan module from EC93 was functional in cell-mediated CDI when fused to the N-terminal portion of the EC93 CdiA protein. Bioinformatic analyses revealed that the genetic organization of CDI systems shares features with rhs (rearrangement hotspot) loci. Rhs proteins also contain polymorphic C-terminal regions (Rhs-CTs), some of which share significant sequence identity with CdiA-CTs. All rhs genes are followed by small ORFs representing possible rhsI immunity genes, and several Rhs systems encode orphan rhs-CT/rhsI modules. Analysis of rhs-CT/rhsI modules from D. dadantii 3937 demonstrated that Rhs-CTs have growth inhibitory activity, which is specifically blocked by cognate RhsI immunity proteins. Together, these results suggest that Rhs plays a role in intercellular competition and that orphan gene modules expand the diversity of toxic activities deployed by both CDI and Rhs systems.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Figure 1. E. coli EC93 contains an orphan cdiA-CT/cdiI module.
The CDI region of E. coli EC93 is depicted, with cdiB, cdiA and cdiI genes shown in yellow, green and purple, respectively. The cdiA coding region upstream of the encoded VENN motif is shown in light green, and the cdiA-CT sequence is shown in dark green. The orphan cdiA-CT fragment (cdiA-CT o1) and orphan cdiI (cdiI o1) are dark green and purple, respectively. The nucleotide sequence of the cdiI - cdiA-CT o1 junction and the predicted reading frames are shown in detail. Sequences similar to transposable element genes are shown in red.
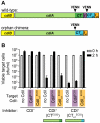
Figure 2. Many bacteria contain orphan cdiA-CT/cdiI modules.
The cdi loci from selected bacterial species are presented using the color-coding scheme in Figure 1. Arrowheads indicate the positions of VENN encoding regions. Probable orphan cdiA-CT/cdiI modules are indicated with an “o” and numeral. Note the change in scale for the locus from E. coli EC869. The UniProt accession numbers for the encoded CdiA proteins are: Escherichia coli 254 ser. O113:H21 (A1XT91); Edwardsiella ictaluri 93–146 (C5BAK2); Erwinia pyrifoliae DSM 12163 (D2T668); Edwardsiella tarda EIB202 (D0ZDG0); Klebsiella variicola At-22 (D3RCW8); Neisseria menigitidis FAM18 (A1KSB8); Photorhabdus asymbiotica ATCC 43949 (B6VKN5); Pectobacterium wasabiae WPP163 (D0KFH4); Yersinia pseudotuberculosis PB1/+ (B2K3A8); and Escherichia coli EC869 (B3BM48). The E. coli 254 genomic sequence does not include the 5′-end of the cdiA gene.
Figure 3. Interchange between cdiA and orphan _cdiA-CT_s.
A) ACT comparison of the CDI systems (region I) from Y. pestis CO92 (top) and Y. pestis Microtus 91001 (bottom). The cdi loci lie within a highly conserved genomic region (red blocks denote nucleotide conservation), but the sequences encoding CdiA-CT diverge beginning at the VENN region. A 3,557 bp deletion in Y. pestis Microtus 91001 has fused the orphan cdiA-CT/cdiI module to the full-length cdiA gene (highlighted in yellow). B) ACT comparison of region II cdi loci from Y. pseudotuberculosis strains IP31758 (top) and PB1/+ (bottom). The cdi loci lie within a highly conserved genomic region (red denotes nucleotide conservation), but the cdiA-CT sequences exhibit complex rearrangements between the two strains. The orphan cdiA-CT o4 PB1(II)/cdiI o4 PB1(II) module of strain PB1/+ is essentially identical to the cdiA-CT IP31758(II) and cdiI IP31758(II) sequences from strain IP31758 (99.1% identity over 3,648 nucleotides; highlighted in yellow). Additionally, the orphan cdiA-CT o1 PB1(II)/cdiI o1 PB1(II) module is nearly identical to the orphan cdiA-CT o2 IP31758(II)/cdiI o2 IP31758(II) module (98% identity over 2,656 nucleotides, highlighted in blue), and the cdiA-CT o2 PB1(II)/cdiI o2 PB1(II) module is nearly identical to a cdiA fragment (and associated cdiI gene) found between the two orphan modules in strain IP31758 (98% identity over 1,073 nucleotides; highlighted in green).
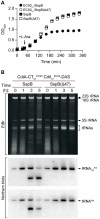
Figure 4. The tRNase activity of CdiA-CTo1 EC93 is blocked by the binding of CdiIo1 EC93.
A) Analysis of CdiA-CT/CdiI binding. Purified CdiA-CT and CdiI-His6 proteins were mixed at equimolar ratios then purified by Ni2+-affinity chromatography. Input samples represent the protein mixtures prior to chromatography. Unbound fractions contain proteins that failed to bind the affinity resin. Bound proteins were eluted from the affinity resin with imidazole. All fractions were analyzed by SDS-PAGE. B) Northern blot analysis of CdiA-CTUPEC536 and CdiA-CTo1 EC93 tRNase activity. S100 fractions containing cellular tRNA was treated with purified CdiA-CT and/or CdiI-His6 proteins and then analyzed by Northern blot hybridization using probes specific for tRNA1B Ala and tRNAHis.
Figure 5. CdiA-CTo1 EC93 inhibits the growth of E. coli cells.
A) Growth curves of E. coli Δ_sspB_ cells expressing CdiA-CTo1 EC93/CdiIo1 EC93-DAS. Degradation of CdiIo1 EC93-DAS was initiated by the addition of L-arabinose to induce SspB synthesis. Control cells express SspB(Δ47), which does not deliver CdiIo1 EC93-DAS to the ClpXP protease. Growth curves with square symbols represent control strains expressing SspB or SspB(Δ47), but not CdiA-CTo1 EC93/CdiIo1 EC93-DAS. B) Analysis of in vivo CdiA-CTo1 EC93 tRNase activity. Total RNA was isolated from cells expressing CdiA-CTo1 EC93/CdiIo1 EC93-DAS at varying times after L-arabinose induction. Samples were run on polyacrylamide gels followed by staining with ethidium bromide (EtBr) or Northern blot analysis using probes specific for tRNA1B Ala and tRNAHis.
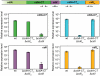
Figure 6. The EC93 orphan cdiA-CT/cdiI module is functional in contact-dependent growth inhibition (CDI).
A) The wild-type EC93 and orphan chimera CDI systems are shown schematically. The cdiA-CT EC93 /cdiI EC93 region was deleted and orphan module fused onto the cdiA EC93 gene at the VENN encoding sequence. B) Growth competitions. CDI+ inhibitor cells were co-cultured with target cells expressing either CdiIEC93 or orphan CdiIo1 EC93 immunity proteins. Viable target cells were quantified by plating on selective media to determine the number of colony forming units (cfu) per milliliter.
Figure 7. The EC93 orphan region is transcribed.
RNA from E. coli EC93, EC93 Δ_cdiA-CT_ EC93Δ_cdiI_ EC93, and EC93 Δ_cdiA-CT_ o1 EC93Δ_cdiI_ o1 EC93 was subjected to quantitative RT-PCR. The primer binding sites within the cdi locus are depicted schematically as arrows. The relative expression levels represent the mean ± SEM for three independently isolated RNA samples.
Figure 8. The EC93 orphan region produces functional CdiIo1 immunity protein.
EC93 expressing chimeric CdiAEC93-CTo1 EC93 was used as an inhibitor strain in growth competition experiments. Inhibitor cells were co-cultured with wild-type EC93, EC93 deleted for the orphan region (Δ_cdiA-CT_ o1 EC93Δ_cdiI_ o1 EC93), and EC93 Δ_cdiA-CT_ o1 EC93Δ_cdiI_ o1 EC93 cells complemented with a plasmid-borne copy of cdiI o1 EC93 (cdiI o1 +). Viable target cells were quantified by plating on selective media to determine the number of colony forming units (cfu) per milliliter.
Figure 9. Rhs genes have variable CT encoding sequences, immunity genes, and orphan rhs-CT/rhsI modules.
ACT comparison of related Rhs regions from Y. pseudotuberculosis strains IP31758 (top) and IP32953 (bottom). This region is highly conserved between the two strains, but the rhs genes encode unrelated CT sequences and the adjacent rhsI genes are unrelated. Both strains contain a related orphan rhs-CT/rhsI module. The orphan module in strain IP32953 has more upstream rhs coding sequence, but contains an in-frame stop codon in this retained sequence. The vgrG gene is a conserved component of Type VI secretion systems; TPR is a conserved gene encoding a potential tetratricopeptide repeat protein; and X and Y are conserved predicted genes of unknown function.
Figure 10. The Rhs genes of D. dadantii 3937 encode toxin/immunity pairs.
E. coli sspB + cells expressing RhsI3937 proteins were incubated with supercoiled plasmids encoding the various Rhs-CT3937/RhsI3937-DAS pairs and plated to select stable transformants. All Rhs-CT3937/RhsI3937-DAS constructs were also introduced into E. coli Δ_sspB_ cells to demonstrate that RhsI3937-DAS degradation is required for growth inhibition.
Similar articles
- A widespread family of polymorphic contact-dependent toxin delivery systems in bacteria.
Aoki SK, Diner EJ, de Roodenbeke CT, Burgess BR, Poole SJ, Braaten BA, Jones AM, Webb JS, Hayes CS, Cotter PA, Low DA. Aoki SK, et al. Nature. 2010 Nov 18;468(7322):439-42. doi: 10.1038/nature09490. Nature. 2010. PMID: 21085179 Free PMC article. - Mechanisms and biological roles of contact-dependent growth inhibition systems.
Hayes CS, Koskiniemi S, Ruhe ZC, Poole SJ, Low DA. Hayes CS, et al. Cold Spring Harb Perspect Med. 2014 Feb 1;4(2):a010025. doi: 10.1101/cshperspect.a010025. Cold Spring Harb Perspect Med. 2014. PMID: 24492845 Free PMC article. Review. - Identification of a contact-dependent growth inhibition system in the probiotic Escherichia coli Nissle 1917.
Chen H, Fang Q, Tu Q, Liu C, Yin J, Yin Y, Xia L, Bian X, Zhang Y. Chen H, et al. FEMS Microbiol Lett. 2018 Jun 1;365(11). doi: 10.1093/femsle/fny102. FEMS Microbiol Lett. 2018. PMID: 29688444 - Diversification of β-Augmentation Interactions between CDI Toxin/Immunity Proteins.
Morse RP, Willett JL, Johnson PM, Zheng J, Credali A, Iniguez A, Nowick JS, Hayes CS, Goulding CW. Morse RP, et al. J Mol Biol. 2015 Nov 20;427(23):3766-84. doi: 10.1016/j.jmb.2015.09.020. Epub 2015 Oct 9. J Mol Biol. 2015. PMID: 26449640 Free PMC article. - Contact-Dependent Growth Inhibition (CDI) and CdiB/CdiA Two-Partner Secretion Proteins.
Willett JL, Ruhe ZC, Goulding CW, Low DA, Hayes CS. Willett JL, et al. J Mol Biol. 2015 Nov 20;427(23):3754-65. doi: 10.1016/j.jmb.2015.09.010. Epub 2015 Sep 24. J Mol Biol. 2015. PMID: 26388411 Free PMC article. Review.
Cited by
- Polymorphic toxin systems: Comprehensive characterization of trafficking modes, processing, mechanisms of action, immunity and ecology using comparative genomics.
Zhang D, de Souza RF, Anantharaman V, Iyer LM, Aravind L. Zhang D, et al. Biol Direct. 2012 Jun 25;7:18. doi: 10.1186/1745-6150-7-18. Biol Direct. 2012. PMID: 22731697 Free PMC article. - A resveratrol oligomer, hopeaphenol suppresses virulence activity of Pectobacterium atrosepticum via the modulation of the master regulator, FlhDC.
Kang JE, Hwang S, Yoo N, Kim BS, Chung EH. Kang JE, et al. Front Microbiol. 2022 Oct 28;13:999522. doi: 10.3389/fmicb.2022.999522. eCollection 2022. Front Microbiol. 2022. PMID: 36386642 Free PMC article. - A new family of secreted toxins in pathogenic Neisseria species.
Jamet A, Jousset AB, Euphrasie D, Mukorako P, Boucharlat A, Ducousso A, Charbit A, Nassif X. Jamet A, et al. PLoS Pathog. 2015 Jan 8;11(1):e1004592. doi: 10.1371/journal.ppat.1004592. eCollection 2015 Jan. PLoS Pathog. 2015. PMID: 25569427 Free PMC article. - Complete genome sequence of Marinomonas posidonica type strain (IVIA-Po-181(T)).
Lucas-Elío P, Goodwin L, Woyke T, Pitluck S, Nolan M, Kyrpides NC, Detter JC, Copeland A, Lu M, Bruce D, Detter C, Tapia R, Han S, Land ML, Ivanova N, Mikhailova N, Johnston AW, Sanchez-Amat A. Lucas-Elío P, et al. Stand Genomic Sci. 2012 Oct 10;7(1):31-43. doi: 10.4056/sigs.2976373. Epub 2012 Sep 27. Stand Genomic Sci. 2012. PMID: 23458837 Free PMC article. - Nice or nasty: Protein translocation between bacteria and the different forms of response.
Unterweger D, Griffin AS. Unterweger D, et al. Proc Natl Acad Sci U S A. 2016 Aug 2;113(31):8559-61. doi: 10.1073/pnas.1609443113. Epub 2016 Jul 15. Proc Natl Acad Sci U S A. 2016. PMID: 27422548 Free PMC article. No abstract available.
References
- Aoki SK, Pamma R, Hernday AD, Bickham JE, Braaten BA, et al. Contact-dependent inhibition of growth in Escherichia coli. Science. 2005;309:1245–1248. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials