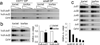A role for cohesin in T-cell-receptor rearrangement and thymocyte differentiation - PubMed (original) (raw)
. 2011 Aug 10;476(7361):467-71.
doi: 10.1038/nature10312.
Bingtao Hao, Kikuë Tachibana-Konwalski, Thais Lavagnolli, Hegias Mira-Bontenbal, Karen E Brown, Grace Teng, Tom Carroll, Anna Terry, Katie Horan, Hendrik Marks, David J Adams, David G Schatz, Luis Aragon, Amanda G Fisher, Michael S Krangel, Kim Nasmyth, Matthias Merkenschlager
Affiliations
- PMID: 21832993
- PMCID: PMC3179485
- DOI: 10.1038/nature10312
A role for cohesin in T-cell-receptor rearrangement and thymocyte differentiation
Vlad C Seitan et al. Nature. 2011.
Abstract
Cohesin enables post-replicative DNA repair and chromosome segregation by holding sister chromatids together from the time of DNA replication in S phase until mitosis. There is growing evidence that cohesin also forms long-range chromosomal cis-interactions and may regulate gene expression in association with CTCF, mediator or tissue-specific transcription factors. Human cohesinopathies such as Cornelia de Lange syndrome are thought to result from impaired non-canonical cohesin functions, but a clear distinction between the cell-division-related and cell-division-independent functions of cohesion--as exemplified in Drosophila--has not been demonstrated in vertebrate systems. To address this, here we deleted the cohesin locus Rad21 in mouse thymocytes at a time in development when these cells stop cycling and rearrange their T-cell receptor (TCR) α locus (Tcra). Rad21-deficient thymocytes had a normal lifespan and retained the ability to differentiate, albeit with reduced efficiency. Loss of Rad21 led to defective chromatin architecture at the Tcra locus, where cohesion-binding sites flank the TEA promoter and the Eα enhancer, and demarcate Tcra from interspersed Tcrd elements and neighbouring housekeeping genes. Cohesin was required for long-range promoter-enhancer interactions, Tcra transcription, H3K4me3 histone modifications that recruit the recombination machinery and Tcra rearrangement. Provision of pre-rearranged TCR transgenes largely rescued thymocyte differentiation, demonstrating that among thousands of potential target genes across the genome, defective Tcra rearrangement was limiting for the differentiation of cohesin-deficient thymocytes. These findings firmly establish a cell-division-independent role for cohesin in Tcra locus rearrangement and provide a comprehensive account of the mechanisms by which cohesin enables cellular differentiation in a well-characterized mammalian system.
Figures
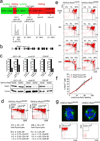
Figure 1. Genetic cohesin depleted in non-dividing thymocytes
a) Thymocyte differentiation from left to right: CD4− CD8− double negative (DN) stages 1 to 4; CD4+ CD8+ double positive (DP), CD4+8lo; CD4 or CD8 single positive (SP) cells. Proliferation is in green, cell cycle arrest in red. Histograms show DNA content. b) Conditional Rad21 allele (see supplementary fig. 1a). c) Real time genomic PCR of Rad21 locus deletion, RT-PCR of Rad21 RNA and western blotting of Rad21 protein. d) Cell numbers and flow cytometric analysis of thymocyte subsets in 6 week-old CD4Cre _Rad21_lox/lox and CD4Cre _Rad21_lox/lwt mice (mean ± SD, n = 12). e) Pulse chase analysis of CD4Cre _Rad21_lox/wt and CD4Cre _Rad21_lox/lox thymocytes. Dot blots are gated on BrdU+ cells (see supplementary fig. 2b). f) Continuous BrdU labelling for DP thymocyte turnover. See supplementary Fig. 2c for CD4+8lo and CD4 SP subsets (mean ± SD, n = 3–5 per data point). g) Top: metaphase spreads of 2 day activated thymocytes stained for alpha-tubulin (green) and DNA (DAPI, blue, see supplementary fig. 3). Bottom: Cells recovered after 5 days.
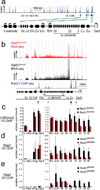
Figure 2. Cohesin affects Tcra transcription Rag recombinase recruitment
a) Rad21 ChIP-seq of the 3' part of the Tcra locus in DP thymocytes. Arrowheads highlight cohesin sites at the Eα enhancer (black), the Tcra locus control region (grey), Jα promoters (turquoise), the TEA promoter (green) and between Tcrd elements and V gene segments (blue). b) RNA-seq of Tcra in CD4Cre _Rad21_lox/lox (red) and control _Rad21_lox/wt (black) DP thymocytes. Rad21 ChIP-seq is in blue. c) ChIP of H3K4me3 relative to total H3 in CD4Cre _Rad21_lox/lox and control DP thymocytes. Hbb is a negative and Actg and Elp4 are positive control loci (mean ± SE of 2 independent experiments). p=0.016 for all Jα elements; p=0.38 (NS) for proximal (Jα61–48) and p=0.004 for distal (Jα37–16) Jα elements. d) ChIP of Rag2 relative to total H3 as in c) (mean ± SE of 3 independent experiments). p=0.0001 for all Jα elements; p=0.054 (NS) for proximal (Jα61–48) and p=0.0001 for distal (Jα37–16) Jα elements. e) ChIP of Rag1 relative to total H3 as in c) (mean ± SE of 2 independent experiments). p=0.005 for all Jα elements; p=0.17 (NS) for proximal (Jα61–48) and p=0.003 for distal (Jα37–16) Jα elements.
Figure 3. Cohesin affects Tcra rearrangement
a) Three-fold dilutions of genomic Vα8-Jα PCR products from CD71+ or CD71− DP thymocytes visualised with ethidium bromide. Cd14 is a genomic control. b) Genomic PCR products from small DP thymocytes visualised by Southern blotting with Jα-specific probes (left). Usage of the distal Jα22 element was reduced by 86% (middle, mean ± SE, n=3). Southern blotting of Vα8-Jα RT-PCR products from CD4Cre _Rad21_lox/lox normalised to control DP thymocytes (right, mean ± SE, n=3). c) Double strand breaks in three-fold serially diluted genomic DNA from CD4Cre Rad21lox/lox and control DP thymocytes detected by ligation-mediated PCR.
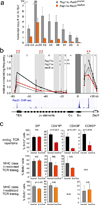
Figure 4. Cohesin mediates long-range interactions between regulatory elements that control Tcra transcription
a) Transcript copy number of the unrearranged Tcra J region in _Rag1_-deficient CD4Cre _Rad21_lox/lox and _Rad21_lox/wt DP thymocytes (mean ± SE, n=3). b) 3C analysis of long-range interactions between Eα and Tcra restriction fragments (shaded) in _Rag1_-deficient CD4Cre _Rad21_lox/wt (black, mean ± SD, n=3), CD4Cre _Rad21_lox/lox (red, mean ± SD, n=3) DP thymocytes and pre-B cells (grey, mean ± SD, n=3). Intervening HindIII fragment numbers and genomic distances are indicated. Stars: p < 0.05 (grey: control thymocytes versus pre-B cells; red: control versus cohesin-depleted thymocytes; NS = not significant). c) TCR transgenes rescue the differentiation of cohesin-depleted thymocytes. Top: Percentages BrdU+ cells in CD4+8lo, CD4 SP and CD8 SP thymocytes. The differentiation of cohesin-deficient thymocytes is rescued by MHC class II-restricted (middle) and MHC class I-restricted TCR transgenes (bottom), n=3–5 per data point ± SD.
Similar articles
- The expanding phenotypes of cohesinopathies: one ring to rule them all!
Piché J, Van Vliet PP, Pucéat M, Andelfinger G. Piché J, et al. Cell Cycle. 2019 Nov;18(21):2828-2848. doi: 10.1080/15384101.2019.1658476. Epub 2019 Sep 13. Cell Cycle. 2019. PMID: 31516082 Free PMC article. Review. - Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus.
Hadjur S, Williams LM, Ryan NK, Cobb BS, Sexton T, Fraser P, Fisher AG, Merkenschlager M. Hadjur S, et al. Nature. 2009 Jul 16;460(7253):410-3. doi: 10.1038/nature08079. Epub 2009 May 20. Nature. 2009. PMID: 19458616 Free PMC article. - Calpain-1 cleaves Rad21 to promote sister chromatid separation.
Panigrahi AK, Zhang N, Mao Q, Pati D. Panigrahi AK, et al. Mol Cell Biol. 2011 Nov;31(21):4335-47. doi: 10.1128/MCB.06075-11. Epub 2011 Aug 29. Mol Cell Biol. 2011. PMID: 21876002 Free PMC article. - CTCF physically links cohesin to chromatin.
Rubio ED, Reiss DJ, Welcsh PL, Disteche CM, Filippova GN, Baliga NS, Aebersold R, Ranish JA, Krumm A. Rubio ED, et al. Proc Natl Acad Sci U S A. 2008 Jun 17;105(24):8309-14. doi: 10.1073/pnas.0801273105. Epub 2008 Jun 11. Proc Natl Acad Sci U S A. 2008. PMID: 18550811 Free PMC article. - Cohesin at active genes: a unifying theme for cohesin and gene expression from model organisms to humans.
Dorsett D, Merkenschlager M. Dorsett D, et al. Curr Opin Cell Biol. 2013 Jun;25(3):327-33. doi: 10.1016/j.ceb.2013.02.003. Epub 2013 Mar 1. Curr Opin Cell Biol. 2013. PMID: 23465542 Free PMC article. Review.
Cited by
- IL-7 coordinates proliferation, differentiation and Tcra recombination during thymocyte β-selection.
Boudil A, Matei IR, Shih HY, Bogdanoski G, Yuan JS, Chang SG, Montpellier B, Kowalski PE, Voisin V, Bashir S, Bader GD, Krangel MS, Guidos CJ. Boudil A, et al. Nat Immunol. 2015 Apr;16(4):397-405. doi: 10.1038/ni.3122. Epub 2015 Mar 2. Nat Immunol. 2015. PMID: 25729925 Free PMC article. - An anti-silencer- and SATB1-dependent chromatin hub regulates Rag1 and Rag2 gene expression during thymocyte development.
Hao B, Naik AK, Watanabe A, Tanaka H, Chen L, Richards HW, Kondo M, Taniuchi I, Kohwi Y, Kohwi-Shigematsu T, Krangel MS. Hao B, et al. J Exp Med. 2015 May 4;212(5):809-24. doi: 10.1084/jem.20142207. Epub 2015 Apr 6. J Exp Med. 2015. PMID: 25847946 Free PMC article. - A role of the CTCF binding site at enhancer Eα in the dynamic chromatin organization of the Tcra-Tcrd locus.
Zhao H, Li Z, Zhu Y, Bian S, Zhang Y, Qin L, Naik AK, He J, Zhang Z, Krangel MS, Hao B. Zhao H, et al. Nucleic Acids Res. 2020 Sep 25;48(17):9621-9636. doi: 10.1093/nar/gkaa711. Nucleic Acids Res. 2020. PMID: 32853367 Free PMC article. - STAG2 loss-of-function mutation induces PD-L1 expression in U2OS cells.
Nie Z, Gao W, Zhang Y, Hou Y, Liu J, Li Z, Xue W, Ye X, Jin A. Nie Z, et al. Ann Transl Med. 2019 Apr;7(7):127. doi: 10.21037/atm.2019.02.23. Ann Transl Med. 2019. PMID: 31157248 Free PMC article. - Genome Topology Control of Antigen Receptor Gene Assembly.
Allyn BM, Lee KD, Bassing CH. Allyn BM, et al. J Immunol. 2020 May 15;204(10):2617-2626. doi: 10.4049/jimmunol.1901356. J Immunol. 2020. PMID: 32366683 Free PMC article. Review.
References
- Nasmyth K, Haering CH. Cohesin: its roles and mechanisms. Annu Rev Genet. 2009;43:525–558. - PubMed
- Hagstrom KA, Meyer BJ. Condensin and cohesin: more than chromosome compactor and glue. Nat Rev Genet. 2003;4:520–534. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- MC_U120027516/MRC_/Medical Research Council/United Kingdom
- R37 GM041052-22/GM/NIGMS NIH HHS/United States
- R37 GM041052/GM/NIGMS NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- 13031/CRUK_/Cancer Research UK/United Kingdom
- MC_U120081295/MRC_/Medical Research Council/United Kingdom
- R37 AI032524-20/AI/NIAID NIH HHS/United States
- R37 AI032524/AI/NIAID NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases