Advances in genetic manipulation of obligate intracellular bacterial pathogens - PubMed (original) (raw)
Advances in genetic manipulation of obligate intracellular bacterial pathogens
Paul A Beare et al. Front Microbiol. 2011.
Abstract
Infections by obligate intracellular bacterial pathogens result in significant morbidity and mortality worldwide. These bacteria include Chlamydia spp., which causes millions of cases of sexually transmitted disease and blinding trachoma annually, and members of the α-proteobacterial genera Anaplasma, Ehrlichia, Orientia, and Rickettsia, agents of serious human illnesses including epidemic typhus. Coxiella burnetii, the agent of human Q fever, has also been considered a prototypical obligate intracellular bacterium, but recent host cell-free (axenic) growth has rescued it from obligatism. The historic genetic intractability of obligate intracellular bacteria has severely limited molecular dissection of their unique lifestyles and virulence factors involved in pathogenesis. Host cell restricted growth is a significant barrier to genetic transformation that can make simple procedures for free-living bacteria, such as cloning, exceedingly difficult. Low transformation efficiency requiring long-term culture in host cells to expand small transformant populations is another obstacle. Despite numerous technical limitations, the last decade has witnessed significant gains in genetic manipulation of obligate intracellular bacteria including allelic exchange. Continued development of genetic tools should soon enable routine mutation and complementation strategies for virulence factor discovery and stimulate renewed interest in these refractory pathogens. In this review, we discuss the technical challenges associated with genetic transformation of obligate intracellular bacteria and highlight advances made with individual genera.
Keywords: allelic exchange; antibiotic selection; complementation; electroporation; genetic transformation; shuttle vector; transposon mutagenesis; virulence factor.
Figures
Figure 1
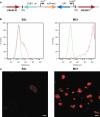
A Himar1 insertion in R. rickettsii sca2 eliminates actin-based motility. The right micrograph shows wild-type R. rickettsii (arrows) with typical filamentous actin tails (arrowhead). The left micrograph depicts a small plaque-forming mutant of R. rickettsii with a Himar1 insertion in sca2, which encodes an autotransporter protein (Kleba et al., 2010). The mutant rickettsia (arrow) lack actin tails. Rickettsia were stained by immunofluorescence using a specific monoclonal antibody and filamentous actin was stained with Alex Fluor 598 phalloidin. Bar, 3 μm. (Micrographs courtesy of Ted Hackstadt, Rocky Mountain Laboratories).
Figure 2
Characterization C. burnetii Nine Mile (phase II) MC1 and B2c Himar1 transformants. (A) Schematic of the Himar1 chromosomal integration site in C. burnetii MC1. The Himar1 transposon is flanked by inverted terminal repeat (ITR) elements and inserted into an intergenic region between CBU0316 and CBU0317. Flow cytometry (B) and confocal fluorescence microscopy (C) of live Vero cells infected with MC1 or B2c for 5 days. Both assays revealed considerably more mCherry fluorescence from MC1-containing vacuoles, where expression is driven by 311P, then from B2c-containing vacuoles, where expression is driven by 1169P (Beare et al., 2009). The green trace in flow cytometry histograms shows autofluorescence of Vero cells infected with wild-type C. burnetii for 5 days. Bars, 5 μm.
Figure 3
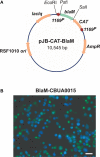
A C. burnetii BlaM translocation assay shuttle vector. (A) Map of the reporter plasmid pJB-CAT-BlaM which contains an RSF1010 ori that functions in both E. coli and C. burnetii. Genes encoding suspected secreted proteins are cloned downstream and in-frame with blaM using the unique SalI site. (B) BlaM translocation assay showing cytosolic delivery of a BlaM-CBUA0015 fusion protein. THP-1 cells were infected with C. burnetii Nine Mile (phase II) containing pJB-CAT-BlaM::CBUA0015 for 48 h, then incubated for 1 h with CCF4/AM. Cleavage of CCF4/AM by cytosolic BlaM results in blue fluorescent cells and indicates secretion of the fusion protein. Bar, 30 μm.
Figure 4
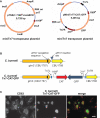
Coxiella burnetii Tn7 transformation system. (A) Maps of two-plasmid C. burnetii miniTn7 transposon system. The suicide plasmid pTNS2_::1169P-tnsABCD_ encodes the tnsABCD operon under control of 1169P. The suicide plasmid pMiniTn7T-CAT::GFP encodes CAT and GFP genes driven independently by 1169P and 311P, respectively. The CAT/GFP cassette is flanked by Tn7L and Tn7R elements. (B) Schematic of the glmS regions in C. burnetii Nine Mile (phase II) and C. burnetii Nine Mile (phase II)/Tn7-CAT-GFP. (C) Fluorescence microscopy of Vero cells infected for 5 days with C. burnetii Nine Mile (phase II)/Tn7-CAT-GFP (green). Cells were fixed with 4% paraformaldehyde, then immunostained for the lysosomal protein CD63 (red). Bar, 5 μm.
Similar articles
- Genetic manipulation of Coxiella burnetii.
Beare PA. Beare PA. Adv Exp Med Biol. 2012;984:249-71. doi: 10.1007/978-94-007-4315-1_13. Adv Exp Med Biol. 2012. PMID: 22711636 Review. - Recent advances in genetic systems in obligate intracellular human-pathogenic bacteria.
Fisher DJ, Beare PA. Fisher DJ, et al. Front Cell Infect Microbiol. 2023 Jun 19;13:1202245. doi: 10.3389/fcimb.2023.1202245. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 37404720 Free PMC article. Review. - Engineering of obligate intracellular bacteria: progress, challenges and paradigms.
McClure EE, Chávez ASO, Shaw DK, Carlyon JA, Ganta RR, Noh SM, Wood DO, Bavoil PM, Brayton KA, Martinez JJ, McBride JW, Valdivia RH, Munderloh UG, Pedra JHF. McClure EE, et al. Nat Rev Microbiol. 2017 Sep;15(9):544-558. doi: 10.1038/nrmicro.2017.59. Epub 2017 Jun 19. Nat Rev Microbiol. 2017. PMID: 28626230 Free PMC article. Review. - Emancipating Chlamydia: Advances in the Genetic Manipulation of a Recalcitrant Intracellular Pathogen.
Bastidas RJ, Valdivia RH. Bastidas RJ, et al. Microbiol Mol Biol Rev. 2016 Mar 30;80(2):411-27. doi: 10.1128/MMBR.00071-15. Print 2016 Jun. Microbiol Mol Biol Rev. 2016. PMID: 27030552 Free PMC article. Review. - Manipulation of Host Cholesterol by Obligate Intracellular Bacteria.
Samanta D, Mulye M, Clemente TM, Justis AV, Gilk SD. Samanta D, et al. Front Cell Infect Microbiol. 2017 May 5;7:165. doi: 10.3389/fcimb.2017.00165. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 28529926 Free PMC article. Review.
Cited by
- Lysosomal degradation products induce Coxiella burnetii virulence.
Newton P, Thomas DR, Reed SCO, Lau N, Xu B, Ong SY, Pasricha S, Madhamshettiwar PB, Edgington-Mitchell LE, Simpson KJ, Roy CR, Newton HJ. Newton P, et al. Proc Natl Acad Sci U S A. 2020 Mar 24;117(12):6801-6810. doi: 10.1073/pnas.1921344117. Epub 2020 Mar 9. Proc Natl Acad Sci U S A. 2020. PMID: 32152125 Free PMC article. - Mosaic composition of ribA and wspB genes flanking the virB8-D4 operon in the Wolbachia supergroup B-strain, wStr.
Baldridge GD, Li YG, Witthuhn BA, Higgins L, Markowski TW, Baldridge AS, Fallon AM. Baldridge GD, et al. Arch Microbiol. 2016 Jan;198(1):53-69. doi: 10.1007/s00203-015-1154-8. Epub 2015 Sep 23. Arch Microbiol. 2016. PMID: 26400107 Free PMC article. - Proteomic analysis of a mosquito host cell response to persistent Wolbachia infection.
Baldridge G, Higgins L, Witthuhn B, Markowski T, Baldridge A, Armien A, Fallon A. Baldridge G, et al. Res Microbiol. 2017 Sep;168(7):609-625. doi: 10.1016/j.resmic.2017.04.005. Epub 2017 Apr 21. Res Microbiol. 2017. PMID: 28435138 Free PMC article. - Critical Role for Molecular Iron in Coxiella burnetii Replication and Viability.
Sanchez SE, Omsland A. Sanchez SE, et al. mSphere. 2020 Jul 22;5(4):e00458-20. doi: 10.1128/mSphere.00458-20. mSphere. 2020. PMID: 32699121 Free PMC article. - Two systems for targeted gene deletion in Coxiella burnetii.
Beare PA, Larson CL, Gilk SD, Heinzen RA. Beare PA, et al. Appl Environ Microbiol. 2012 Jul;78(13):4580-9. doi: 10.1128/AEM.00881-12. Epub 2012 Apr 20. Appl Environ Microbiol. 2012. PMID: 22522687 Free PMC article.
References
- Baldridge G. D., Burkhardt N., Herron M. J., Kurtti T. J., Munderloh U. G. (2005). Analysis of fluorescent protein expression in transformants of Rickettsia monacensis, an obligate intracellular tick symbiont. Appl. Environ. Microbiol. 71, 2095–210510.1128/AEM.71.4.2095-2105.2005 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous