TopHat-Fusion: an algorithm for discovery of novel fusion transcripts - PubMed (original) (raw)
TopHat-Fusion: an algorithm for discovery of novel fusion transcripts
Daehwan Kim et al. Genome Biol. 2011.
Abstract
TopHat-Fusion is an algorithm designed to discover transcripts representing fusion gene products, which result from the breakage and re-joining of two different chromosomes, or from rearrangements within a chromosome. TopHat-Fusion is an enhanced version of TopHat, an efficient program that aligns RNA-seq reads without relying on existing annotation. Because it is independent of gene annotation, TopHat-Fusion can discover fusion products deriving from known genes, unknown genes and unannotated splice variants of known genes. Using RNA-seq data from breast and prostate cancer cell lines, we detected both previously reported and novel fusions with solid supporting evidence. TopHat-Fusion is available at http://tophat-fusion.sourceforge.net/.
Figures
Figure 1
Read distributions around two fusions: _BCAS4_-BCAS3 and _TOB1_-SYNRG. (a) Sixty reads aligned by TopHat-Fusion that identify a fusion product formed by the BCAS4 gene on chromosome 20 and the BCAS3 gene on chromosome 17. The data contained more reads than shown; they are collapsed to illustrate how well they are distributed. The inset figures show the coverage depth in 600-bp windows around each fusion. (b) TOB1 (ENSG00000141232)-SYNRG is a novel fusion gene found by TopHat-Fusion, shown here with 70 reads mapping across the fusion point. Note that some of the reads in green span an intron (indicated by thin horizontal lines extending to the right), a feature that can be detected by TopHat's spliced alignment procedure.
Figure 2
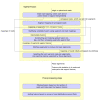
TopHat-Fusion pipeline. TopHat-Fusion consists of two main modules: (1) finding candidate fusions and aligning reads across them; and (2) filtering out false fusions using a series of post-processing routines.
Figure 3
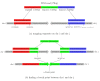
Aligning a read that spans a fusion point. (a) An initially unmapped read of 75 bp is split into three segments of 25 bp, each of which is mapped separately. As shown here, the left (red) and right (blue) segments are mapped to two different chromosomes, i and j. (b) The unmapped green segment is used to find the precise fusion point between i and j. This is done by aligning the green segment to the sequences just to the right of the red segment on chromosome i and just to the left of the blue segment on chromosome j.
Figure 4
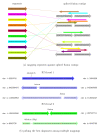
Mapping against fusion points and selecting best read alignments. (a) Bowtie is used to align all segments from the initially unmapped (IUM) reads against spliced fusion contigs, shown in gray on the right. For example, the brown read on the top left aligns to the first spliced fusion contig on the top right. (b) IUM reads 1 and 2 each have multiple alignments. Read 1 has a gap-free alignment, shown in dark blue, which is preferred over the other two alignments shown in lighter shades of blue. The gap-free alignment with three mismatches is preferred over the fusion alignment with one mismatch. If all alignments have gaps and mismatches, then the algorithm prefers those with fewer mismatches, as shown by the dark green alignment for IUM read 2. Full details of the scoring function that determines these preferences are described in the Materials and methods.
Figure 5
Supporting and contradicting evidence for fusion transcripts. (a) Given a fusion point and the chromosomes (gray) spanning it, single-end and paired-end reads (blue) support the fusion. Other reads (red) contradict the fusion by mapping entirely to either of the two chromosomes. (b) TopHat-Fusion prefers reads that uniformly cover a 600-bp window centered in any fusion point. On the upper left, blue reads cover the entire window. On the lower left, red reads cover only a narrow window around the fusion. On the lower right, reads do not cover part of the 600-bp window. The cases shown in orange will be rejected by TopHat-Fusion.
Figure 6
TopHat-Fusion's scoring scheme of read distributions. A scoring scheme of how well distributed reads are around a fusion point; these result scores are used to sort the list of candidate fusions. Variables are defined in the main text.
Similar articles
- TopHat: discovering splice junctions with RNA-Seq.
Trapnell C, Pachter L, Salzberg SL. Trapnell C, et al. Bioinformatics. 2009 May 1;25(9):1105-11. doi: 10.1093/bioinformatics/btp120. Epub 2009 Mar 16. Bioinformatics. 2009. PMID: 19289445 Free PMC article. - State of art fusion-finder algorithms are suitable to detect transcription-induced chimeras in normal tissues?
Carrara M, Beccuti M, Cavallo F, Donatelli S, Lazzarato F, Cordero F, Calogero RA. Carrara M, et al. BMC Bioinformatics. 2013;14 Suppl 7(Suppl 7):S2. doi: 10.1186/1471-2105-14-S7-S2. Epub 2013 Apr 22. BMC Bioinformatics. 2013. PMID: 23815381 Free PMC article. - Identification of gene fusion transcripts by transcriptome sequencing in BRCA1-mutated breast cancers and cell lines.
Ha KC, Lalonde E, Li L, Cavallone L, Natrajan R, Lambros MB, Mitsopoulos C, Hakas J, Kozarewa I, Fenwick K, Lord CJ, Ashworth A, Vincent-Salomon A, Basik M, Reis-Filho JS, Majewski J, Foulkes WD. Ha KC, et al. BMC Med Genomics. 2011 Oct 27;4:75. doi: 10.1186/1755-8794-4-75. BMC Med Genomics. 2011. PMID: 22032724 Free PMC article. - FusionQ: a novel approach for gene fusion detection and quantification from paired-end RNA-Seq.
Liu C, Ma J, Chang CJ, Zhou X. Liu C, et al. BMC Bioinformatics. 2013 Jun 15;14:193. doi: 10.1186/1471-2105-14-193. BMC Bioinformatics. 2013. PMID: 23768108 Free PMC article. - Discovery and Classification of Fusion Transcripts in Prostate Cancer and Normal Prostate Tissue.
Luo JH, Liu S, Zuo ZH, Chen R, Tseng GC, Yu YP. Luo JH, et al. Am J Pathol. 2015 Jul;185(7):1834-45. doi: 10.1016/j.ajpath.2015.03.008. Epub 2015 May 9. Am J Pathol. 2015. PMID: 25963990 Free PMC article. Review.
Cited by
- Unraveling Gene Fusions for Drug Repositioning in High-Risk Neuroblastoma.
Liu Z, Chen X, Roberts R, Huang R, Mikailov M, Tong W. Liu Z, et al. Front Pharmacol. 2021 Apr 23;12:608778. doi: 10.3389/fphar.2021.608778. eCollection 2021. Front Pharmacol. 2021. PMID: 33967751 Free PMC article. - Integrative analysis of synovial sarcoma transcriptome reveals different types of transcriptomic changes.
Sun Z, Yin M, Ding Y, Zhu Z, Sun Y, Li K, Yan W. Sun Z, et al. Front Genet. 2022 Sep 2;13:925564. doi: 10.3389/fgene.2022.925564. eCollection 2022. Front Genet. 2022. PMID: 36118864 Free PMC article. - N6-methyladenosine modified circPAK2 promotes lymph node metastasis via targeting IGF2BPs/VEGFA signaling in gastric cancer.
Ding P, Wu H, Wu J, Li T, He J, Ju Y, Liu Y, Li F, Deng H, Gu R, Zhang L, Guo H, Tian Y, Yang P, Meng N, Li X, Guo Z, Meng L, Zhao Q. Ding P, et al. Oncogene. 2024 Aug;43(34):2548-2563. doi: 10.1038/s41388-024-03099-w. Epub 2024 Jul 17. Oncogene. 2024. PMID: 39014193 - New somatic mutations and WNK1-B4GALNT3 gene fusion in papillary thyroid carcinoma.
Costa V, Esposito R, Ziviello C, Sepe R, Bim LV, Cacciola NA, Decaussin-Petrucci M, Pallante P, Fusco A, Ciccodicola A. Costa V, et al. Oncotarget. 2015 May 10;6(13):11242-51. doi: 10.18632/oncotarget.3593. Oncotarget. 2015. PMID: 25803323 Free PMC article. - Integrated sequence and expression analysis of ovarian cancer structural variants underscores the importance of gene fusion regulation.
Mittal VK, McDonald JF. Mittal VK, et al. BMC Med Genomics. 2015 Jul 17;8:40. doi: 10.1186/s12920-015-0118-9. BMC Med Genomics. 2015. PMID: 26177635 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
- R01 HG006677/HG/NHGRI NIH HHS/United States
- R01-HG006102/HG/NHGRI NIH HHS/United States
- R01 HG006102-02/HG/NHGRI NIH HHS/United States
- R01 HG006677-12/HG/NHGRI NIH HHS/United States
- R01-LM006845/LM/NLM NIH HHS/United States
- R01 HG006102/HG/NHGRI NIH HHS/United States
- R01 HG006677-13/HG/NHGRI NIH HHS/United States
- R01 GM083873/GM/NIGMS NIH HHS/United States
- R01 HG006102-01/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources