Structural analysis of the Ras-like G protein MglA and its cognate GAP MglB and implications for bacterial polarity - PubMed (original) (raw)
Structural analysis of the Ras-like G protein MglA and its cognate GAP MglB and implications for bacterial polarity
Mandy Miertzschke et al. EMBO J. 2011.
Abstract
The bacterium Myxococcus xanthus uses a G protein cycle to dynamically regulate the leading/lagging pole polarity axis. The G protein MglA is regulated by its GTPase-activating protein (GAP) MglB, thus resembling Ras family proteins. Here, we show structurally and biochemically that MglA undergoes a dramatic, GDP-GTP-dependent conformational change involving a screw-type forward movement of the central β2-strand, never observed in any other G protein. This movement and complex formation with MglB repositions the conserved residues Arg53 and Gln82 into the active site. Residues required for catalysis are thus not provided by the GAP MglB, but by MglA itself. MglB is a Roadblock/LC7 protein and functions as a dimer to stimulate GTP hydrolysis in a 2:1 complex with MglA. In vivo analyses demonstrate that hydrolysis mutants abrogate Myxococcus' ability to regulate its polarity axis changing the reversal behaviour from stochastic to oscillatory and that both MglA GTPase activity and MglB GAP catalysis are essential for maintaining a proper polarity axis.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
Figure 1
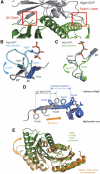
Structure of MglA. (A) Alignment of MglA proteins from M. xanthus (M.x.) and T. thermophilus (T.t.) to Ras-like G proteins from Homo sapiens (H.s.) and Sar1 from Saccharomyces cerivisiae (S.c.). Conserved residues are highlighted in dark and light grey dependent on their degree of conservation. The G1–G5 motifs and switch regions characteristic for the G domain and the secondary structure elements of MglA are shown below and above the alignment, respectively. Red arrows indicate residues mutated for biochemical studies. (B) Structure of _T.t._MglA bound to GDP. Switch I (light blue), switch II (green), P-loop (red) and other characteristic structural elements such as the β2-screw (orange) are indicated. (C) Structures of GDP-bound human H-Ras (2cld), Ran (3GJ0) with switch I β-sheet (blue) and Arl3 (1FZQ) with interswitch toggle (orange).
Figure 2
Structure of MglB. (A) Alignment of bacterial MglB from M. xanthus (M.x.), T. thermophilus (T.t.), Deinococcus geothermalis (D.g.) and Stigmatella aurantiaca (S.a.), with examples of the closest structural homologues, MP1 and p14, of Mus musculus (M.m.) and Xenopus laevis (X.l.). Conserved residues are highlighted in dark and light grey dependent on their degree of conservation. The secondary structure of MglB is indicated above the alignment. Red arrows show residues mutated for biochemical studies and crystallization purposes; blue arrows mark residues mutated without any effect on MglA binding, GAP activity or crystallization. (B) Homodimer of MgB with monomers (Mon) A (green) and B (red). The two-helix surface (left) and four-helix side (right) are related by 180°. (C) Heterodimer of Robl/LC7 domain proteins MP1 (red) and p14 (green) from M. musculus (1VEU). The two-helix side is shown.
Figure 3
The MglA·GppNHp·MglB complex. (A) Structure of MglA·GppNHp (blue) bound to the MglBA5 dimer (green/dark green). Flexible loops that were not visible in electron density are shown with dotted lines. (B) Active site titration. In all, 20 μM of MglA·mant-GppNHp were titrated with increasing amounts of MglB (_K_d of 2 μM) at 37°C in Buffer M and the polarization increase was monitored. (C, D) Analytical size exclusion chromatography (Superdex 75 10/300 GL). (C) Elution profiles of MglB (light green), full-length MglB (dark green) and MglA bound to GDP (light blue) and GppNHp (dark blue). (D) MglA/MglB complex formation is monitored by mixing MglB with MglA·GppNHp (red) and MglA·GDP in presence (orange) or absence (brown) of AlFx as indicated.
Figure 4
Conformational changes and the GDP–GTP structural transition. (A) Superimposition of MglA·GDP (grey) onto the MglA·GppNHp·MglBA5 structure shows how β0 and switch I would clash (red squares). (B) Structural change of switch I on the MglA·GDP (grey) to MglA·GppNHp (light blue) transition, highlighting Arg53 und Thr54. (C) Structural change of switch II on the MglA·GDP (grey) to MglA·GppNHp (light green) transition highlighting Gln82. (D) The β2-screw back-to-front (towards the nucleotide) movement of MglA on the GDP (grey) to GppNHp (blue) transition, reregistering Phe56, Phe57 and Phe59 besides other residues. (E) Structural changes of one MglB protomer in non-complexed MglB (light orange/orange), which bends its α2 side more towards MglA on complex formation (green/dark green).
Figure 5
The hydrophobic interface between MglA·GppNHp and MglB. (A) Residues involved in interface between MglA (blue) and MglB Protomer A and B (green A, B) are schematically indicated. Hydrophobic and Van-der-Waals interactions (solid lines), salt bridges (red dotted lines) and H-bonds (black dotted lines) are shown. (B) Dissociation constants (_K_d) determined by fluorescence polarization during titration of 1 μM MglA·mant-GppNHp with MglB, MglBA68/72R and MglBA5 at 37°C in Buffer M. One representative of three independently carried out experiments is shown. _K_d's and error rates are the ones obtained by the fitting algorithm for the data shown. (C) Kinetics of GTP hydrolysis measured by Pi release from [γ-32P]GTP by the Charcoal Assay at RT in Buffer M. Single turnover conditions were employed with 4 μM nucleotide-free MglA incubated with 1 μM GTP and 40 μM MglB, 40 μM MglBA68/72R or 240 μM MglBA5 thereby ensuring full complex formation. Data were plotted by showing the ratio of specific counts per minute of the supernatant over total counts per minute of sample at each time point. Hydrolysis rates (_k_cat) were obtained by fitting data points to a first-order reaction using Grafit5 (Erithacus software).
Figure 6
A new type of catalytic mechanism. (A) Structure of the MglA·GDP·AlF4−·MglBA5 complex with MglA·GDP in yellow and MglBA5 in green. Flexible loops not visible in the electron density are shown with dotted lines. Zoom into the active site of MglA·GDP·AlF4−·MglBA5 (yellow) superimposed on MglA·GppNHp·MglBA5 (blue). (B, C) Details of the active site of MglA·GppNHp·MglBA5 (B) and comparison to MglA·GDP·AlF4−·MglBA5 (C) Important residues, Thr26, Thr54, Gly81, Gln82 and Arg53, the catalytic water (blue dot) and distances in Angstroms (Å) are indicated.
Figure 7
Mutational studies of the catalytic mechanism. (A) Dissociation constants (_K_d) determined by fluorescence polarization by titrating 1 μM MglAWT, MglAG21V, MglAQ82A and MglAR53A bound to mant-GppNHp with MglB at 37°C in Buffer M. One representative of three independently carried out experiments is shown. _K_d's and error rates, which are shown below, are the ones obtained by the fitting algorithm for the data shown. (B) Intrinsic hydrolysis of different mutants of MglA as described in Figure 5C. (C) GAP-stimulated GTP hydrolysis of MglAWT, MglAG21V, MglAQ82A and MglAR53A measured as described in Figure 5C. Single turnover conditions where 4 μM nucleotide-free MglA proteins were incubated with 1 μM GTP and equimolar amounts of GAP at RT in Buffer M.
Figure 8
MglA GTPase activity and MglB GAP activity are essential for correct localization. Strains of the indicated genotypes were transferred from exponentially growing cultures to a thin agar-pad on a microscope slide, and imaged by time-lapse fluorescence microscopy. Red and blue arrows indicate opposite directions of movement. White arrowheads indicate the oscillating cluster generated by the three mutant MglA proteins.
Similar articles
- A bacterial Ras-like small GTP-binding protein and its cognate GAP establish a dynamic spatial polarity axis to control directed motility.
Zhang Y, Franco M, Ducret A, Mignot T. Zhang Y, et al. PLoS Biol. 2010 Jul 20;8(7):e1000430. doi: 10.1371/journal.pbio.1000430. PLoS Biol. 2010. PMID: 20652021 Free PMC article. - Dual specificity of a prokaryotic GTPase-activating protein (GAP) to two small Ras-like GTPases in Myxococcus xanthus.
Kanade M, Singh NB, Lagad S, Baranwal J, Gayathri P. Kanade M, et al. FEBS J. 2021 Mar;288(5):1565-1585. doi: 10.1111/febs.15513. Epub 2020 Sep 3. FEBS J. 2021. PMID: 32772462 - A dynamic response regulator protein modulates G-protein-dependent polarity in the bacterium Myxococcus xanthus.
Zhang Y, Guzzo M, Ducret A, Li YZ, Mignot T. Zhang Y, et al. PLoS Genet. 2012;8(8):e1002872. doi: 10.1371/journal.pgen.1002872. Epub 2012 Aug 16. PLoS Genet. 2012. PMID: 22916026 Free PMC article. - Unorthodox regulation of the MglA Ras-like GTPase controlling polarity in Myxococcus xanthus.
Dinet C, Mignot T. Dinet C, et al. FEBS Lett. 2023 Mar;597(6):850-864. doi: 10.1002/1873-3468.14565. Epub 2023 Jan 4. FEBS Lett. 2023. PMID: 36520515 Review. - GTPases in bacterial cell polarity and signalling.
Bulyha I, Hot E, Huntley S, Søgaard-Andersen L. Bulyha I, et al. Curr Opin Microbiol. 2011 Dec;14(6):726-33. doi: 10.1016/j.mib.2011.09.001. Epub 2011 Sep 28. Curr Opin Microbiol. 2011. PMID: 21955886 Review.
Cited by
- Crystal structure of the Ego1-Ego2-Ego3 complex and its role in promoting Rag GTPase-dependent TORC1 signaling.
Powis K, Zhang T, Panchaud N, Wang R, De Virgilio C, Ding J. Powis K, et al. Cell Res. 2015 Sep;25(9):1043-59. doi: 10.1038/cr.2015.86. Epub 2015 Jul 24. Cell Res. 2015. PMID: 26206314 Free PMC article. - PomX, a ParA/MinD ATPase activating protein, is a triple regulator of cell division in Myxococcus xanthus.
Schumacher D, Harms A, Bergeler S, Frey E, Søgaard-Andersen L. Schumacher D, et al. Elife. 2021 Mar 18;10:e66160. doi: 10.7554/eLife.66160. Elife. 2021. PMID: 33734087 Free PMC article. - Protein-protein interaction network controlling establishment and maintenance of switchable cell polarity.
Carreira LAM, Tostevin F, Gerland U, Søgaard-Andersen L. Carreira LAM, et al. PLoS Genet. 2020 Jun 22;16(6):e1008877. doi: 10.1371/journal.pgen.1008877. eCollection 2020 Jun. PLoS Genet. 2020. PMID: 32569324 Free PMC article. - Review series: Rab GTPases and membrane identity: causal or inconsequential?
Barr FA. Barr FA. J Cell Biol. 2013 Jul 22;202(2):191-9. doi: 10.1083/jcb.201306010. J Cell Biol. 2013. PMID: 23878272 Free PMC article. Review. - A bipartite, low-affinity roadblock domain-containing GAP complex regulates bacterial front-rear polarity.
Szadkowski D, Carreira LAM, Søgaard-Andersen L. Szadkowski D, et al. PLoS Genet. 2022 Sep 6;18(9):e1010384. doi: 10.1371/journal.pgen.1010384. eCollection 2022 Sep. PLoS Genet. 2022. PMID: 36067225 Free PMC article.
References
- Alto NM (2008) Mimicking small G-proteins: an emerging theme from the bacterial virulence arsenal. Cell Microbiol 10: 566–575 - PubMed
- Bergfors T (2003) Seeds to crystals. J Struct Biol 142: 66–76 - PubMed
- Bos JL, Rehmann H, Wittinghofer A (2007) GEFs and GAPs: critical elements in the control of small G proteins. Cell 129: 865–877 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous