The emergence of genome-based drug repositioning - PubMed (original) (raw)
Comment
The emergence of genome-based drug repositioning
Yves A Lussier et al. Sci Transl Med. 2011.
Abstract
In a pair of papers in this issue of Science Translational Medicine, Butte et al. provide a concrete example of how reinterpreting and comparing genome-wide metrics allows us to effectively hypothesize which drugs from one disease-indication can be repurposed for another disease. The shift toward integrative genome-wide computational approaches has precedence in insightful scalar theories of biological information. Here, we discuss how this recent work in drug repurposing adheres to and takes advantage of the concepts surrounding this information paradigm.
Figures
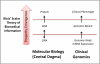
Figure 1. Scalar information flow in molecular biology and present day genomics
The central dogma of molecular biology was explicitly coined by Crick as an information model and is encompassed by a Blois' broader subsequent theory of biomedical information. These scalar theories remain salient to genome-based therapy as they appropriately frame the information intermediaries between molecular mechanisms and clinical-level phenotypes. Above, the flow of information from lower scales to higher ones is shown for the central dogma and for gene expression data. Conventional gene expression signature classifiers predictive of therapeutic utility correspond to an intermediate emergent property, measured by a genomic metric, consisting of a selected number of genes. In contrast, Butte proposes that a drug is a potential candidate for a disease treatment when their respective global genomic metrics are in opposition.
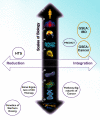
Figure 2. Biological Scales Contrasting Reductionist and Integrative Drug Repositioning Computations
A few exemplar bioinformatics and computational biology studies of drug targeting or repositioning are used to illustrate the major differences in approaches: (i) reduction to the molecular function on the left quadrants, and (ii) integrated properties of multiple molecules to the right. The vertical axis discriminates among different scales of biological substrates for genetic or genomic assays. As shown at the DNA base pair scale, the genetics of warfarin dosage elucidated from a genome-wide association study is illustrated on the lower left quadrant (18). Gene expression signature classifiers follow at the mRNA scale (
). Integrated pathway-level scores of gene expression measures are shown in the lower right quadrant (19). High throughput screens (HTS) (11) of the upper left quadrant correspond to reductionism methods at the tissue level and contrast with increasingly integrative genome-wide tissular methods of the PREDICT algorithm (13) and that of Butte's research group conducted at the same biological scale (upper right quadrant, orange circles). Importantly, the genome-wide metric developed by Butte's group was applied to both a cellular-level disease (cancer) and a tissular one (IBD). As shown by the void in the uppermost part of the right quadrant, systemic diseases and multi-organ ones have not yet been explored by genome-wide metric and may require more comprehensive analyses than straightforward opposition of a genomic metric measured in surrogate tissues and drugs. Scales of biology (m): DNA base pairs, DNA, protein, organelle (mitochondria), cell, tissue, organ, system, organism.
Comment on
- Computational repositioning of the anticonvulsant topiramate for inflammatory bowel disease.
Dudley JT, Sirota M, Shenoy M, Pai RK, Roedder S, Chiang AP, Morgan AA, Sarwal MM, Pasricha PJ, Butte AJ. Dudley JT, et al. Sci Transl Med. 2011 Aug 17;3(96):96ra76. doi: 10.1126/scitranslmed.3002648. Sci Transl Med. 2011. PMID: 21849664 Free PMC article. - Discovery and preclinical validation of drug indications using compendia of public gene expression data.
Sirota M, Dudley JT, Kim J, Chiang AP, Morgan AA, Sweet-Cordero A, Sage J, Butte AJ. Sirota M, et al. Sci Transl Med. 2011 Aug 17;3(96):96ra77. doi: 10.1126/scitranslmed.3001318. Sci Transl Med. 2011. PMID: 21849665 Free PMC article.
Similar articles
- Computational repositioning of the anticonvulsant topiramate for inflammatory bowel disease.
Dudley JT, Sirota M, Shenoy M, Pai RK, Roedder S, Chiang AP, Morgan AA, Sarwal MM, Pasricha PJ, Butte AJ. Dudley JT, et al. Sci Transl Med. 2011 Aug 17;3(96):96ra76. doi: 10.1126/scitranslmed.3002648. Sci Transl Med. 2011. PMID: 21849664 Free PMC article. - Integrative cancer pharmacogenomics to establish drug mechanism of action: drug repurposing.
El-Hachem N, Ba-Alawi W, Smith I, Mer AS, Haibe-Kains B. El-Hachem N, et al. Pharmacogenomics. 2017 Nov;18(16):1469-1472. doi: 10.2217/pgs-2017-0132. Epub 2017 Oct 23. Pharmacogenomics. 2017. PMID: 29057710 No abstract available. - Computational biology approaches for drug repurposing.
Waseem T, Rajput TA, Mushtaq MS, Babar MM, Rajadas J. Waseem T, et al. Prog Mol Biol Transl Sci. 2024;205:91-109. doi: 10.1016/bs.pmbts.2024.03.018. Epub 2024 Apr 4. Prog Mol Biol Transl Sci. 2024. PMID: 38789189 Review. - Computational drug repositioning: from data to therapeutics.
Hurle MR, Yang L, Xie Q, Rajpal DK, Sanseau P, Agarwal P. Hurle MR, et al. Clin Pharmacol Ther. 2013 Apr;93(4):335-41. doi: 10.1038/clpt.2013.1. Epub 2013 Jan 15. Clin Pharmacol Ther. 2013. PMID: 23443757 Review. - Systems biology based drug repositioning for development of cancer therapy.
Turanli B, Altay O, Borén J, Turkez H, Nielsen J, Uhlen M, Arga KY, Mardinoglu A. Turanli B, et al. Semin Cancer Biol. 2021 Jan;68:47-58. doi: 10.1016/j.semcancer.2019.09.020. Epub 2019 Sep 27. Semin Cancer Biol. 2021. PMID: 31568815 Review.
Cited by
- The Omics Revolution Continues: The Maturation of High-Throughput Biological Data Sources.
Mathé E, Hays JL, Stover DG, Chen JL. Mathé E, et al. Yearb Med Inform. 2018 Aug;27(1):211-222. doi: 10.1055/s-0038-1667085. Epub 2018 Aug 29. Yearb Med Inform. 2018. PMID: 30157526 Free PMC article. Review. - Drug-target interaction prediction by integrating chemical, genomic, functional and pharmacological data.
Yang F, Xu J, Zeng J. Yang F, et al. Pac Symp Biocomput. 2014:148-59. Pac Symp Biocomput. 2014. PMID: 24297542 Free PMC article. - A computational method for drug repositioning using publicly available gene expression data.
Shabana KM, Abdul Nazeer KA, Pradhan M, Palakal M. Shabana KM, et al. BMC Bioinformatics. 2015;16 Suppl 17(Suppl 17):S5. doi: 10.1186/1471-2105-16-S17-S5. Epub 2015 Dec 7. BMC Bioinformatics. 2015. PMID: 26679199 Free PMC article. - Translational bioinformatics embraces big data.
Shah NH. Shah NH. Yearb Med Inform. 2012;7(1):130-4. Yearb Med Inform. 2012. PMID: 22890354 Free PMC article. Review. - Translating cancer 'omics' to improved outcomes.
Vucic EA, Thu KL, Robison K, Rybaczyk LA, Chari R, Alvarez CE, Lam WL. Vucic EA, et al. Genome Res. 2012 Feb;22(2):188-95. doi: 10.1101/gr.124354.111. Genome Res. 2012. PMID: 22301133 Free PMC article.
References
- Boguski MS, Mandl KD, Sukhatme VP. Drug discovery. Repurposing with a difference. Science. 2009;324:1394–1395. - PubMed
- Crick F. Central dogma of molecular biology. Nature. 1970;227:561–563. - PubMed
- Blois MS. Information and medicine : the nature of medical descriptions. University of California Press; Berkeley: 1984.
- Simon R. Roadmap for developing and validating therapeutically relevant genomic classifiers. J Clin Oncol. 2005;23:7332–7341. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 3UL1RR024999-03S3/RR/NCRR NIH HHS/United States
- 5UL1RR024999/RR/NCRR NIH HHS/United States
- 5P30CA014599-35/CA/NCI NIH HHS/United States
- UL1 RR024999/RR/NCRR NIH HHS/United States
- U19GM061393-11/GM/NIGMS NIH HHS/United States
- K22 LM008308/LM/NLM NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources