Understanding the transcriptome through RNA structure - PubMed (original) (raw)
Review
Understanding the transcriptome through RNA structure
Yue Wan et al. Nat Rev Genet. 2011.
Abstract
RNA structure is crucial for gene regulation and function. In the past, transcriptomes have largely been parsed by primary sequences and expression levels, but it is now becoming feasible to annotate and compare transcriptomes based on RNA structure. In addition to computational prediction methods, the recent advent of experimental techniques to probe RNA structure by high-throughput sequencing has enabled genome-wide measurements of RNA structure and has provided the first picture of the structural organization of a eukaryotic transcriptome - the 'RNA structurome'. With additional advances in method refinement and interpretation, structural views of the transcriptome should help to identify and validate regulatory RNA motifs that are involved in diverse cellular processes and thereby increase understanding of RNA function.
Figures
Figure 1
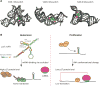
Diversity and dynamics of RNA structures. A, Different classes of SAM riboswitches bind specifically to SAM. The backbone of the riboswitch is in grey while the SAM molecule is colored. SAM-I [PDB number: 2GIS]; SAM-II [PDB number:2QWY]; S(MK) riboswitch [PDB number3E5C]. Images are generated using Pymol. B, Dynamic changes in p27 mRNA structure upon Puf binding results in changes in p27 gene expression. Left panel: During quiescence, the miRNA binding site in 3′UTR of p27 is in a folded structure and is not accessible to miRNA. Translation of p27 mRNA results in high p27 protein levels to maintain quiescence. Right panel: During cellular proliferation, binding of Puf proteins to p27 mRNA causes a structural change that allows miRNA binding sites to be accessible to miR-221 and miR-222, resulting in translation repression of p27. Low p27 levels allow the cells to exit cellular quiescence and enter the cell cycle. [Figure modified from Kedde M. et al, 2010]
Figure 2
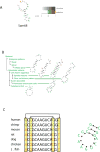
Predicting structural motifs for RNA binding protein (RPB) targets in mRNAs from different organisms. A, By applying an RNA motif finder, investigators were able to identify a significant stem-loop structural motif by analyzing the eight known targets of the human RBP Sam68. B, The same RNA motif finder was used to analyze data collected from a large study on mRNA localization during fly embryonic development, to predict significant motifs in six sets of colocalized maternal transcripts. Shown is the structural motif enriched in each set of mRNAs. C, Conservation of base pairs in homologous sequences directs structure prediction. Sequence covariation found at aligned positions. Shown is an example alignment of seven RNA sequences. In the example, sequence covariation in between the two sets of marked columns hint at interacting bases.
Figure 3
Structure probing by RNA footprinting followed by gel and capillary electrophoresis. An RNA of interest is typically in-vitro transcribed, folded and subjected to a combination of single and double stranded structural probes in solution. Cleavages in double or single stranded regions can either be identified by running a gel electrophoresis (RNA needs to be radioactively labelled at one end) or be identified via primer extension followed by capillary electrophoresis (primer needs to be florescently labelled). The bands from gel electrophoresis can be quantitated using a program called SAFA, while bands in capillary electrophoresis are identified and quantitated using CAFA or ShapeFinder. In gel electrophoresis, the SAFA quantitated green lines refer to the intensity of S1 nuclease cleavages while the red lines refer to the intensity of RNase V1 cleavages. The positions of these cleaved bases are determined from the RNase T1 ladder and alkaline hydrolysis ladder. In capillary electrophoresis, the red line indicates the intensity of structure probing sites that are detected by reverse transcription, while the grey line corresponds to a ladder that positions the RNA bases.
Figure 4
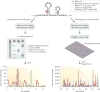
PARS and Frag-seq methods. A, PARS strategy. In PARS, polyA selected RNA is folded in vitro and incubated with either RNase V1 or S1 nuclease to probe for double and single stranded regions respectively. RNase V1 and S1 nuclease cleave resulting in a 5′P leaving group. The enzymatically probed RNA is then fragmented. As enzymatic cleavage products contain 5′P whereas fragmentation and degradation products have 5′OH, only true structure probing sites can be ligated to adapters and reverse transcribed. The cDNA library is sequenced using high throughput sequencing and the resulting reads are mapped to the genome to identify double/single stranded regions in the transcriptome. A PARS score can be calculated at each base whereby a positive PARS score indicates that a base is double stranded and a negative PARS score indicates that a base is single stranded. B, Fragseq stratgy. Nuclear RNA is folded in vitro and probed in solution with P1 endonuclease. P1 cleaves at single stranded regions, resulting in a 5′P leaving group. This 5′P can be captured by adapter ligation, followed by reverse transcription and high throughput sequencing. Sequencing reads are mapped back to the genome to identify where single stranded bases are located in the transcriptome. Fragseq also contains controls which include sequencing of endogenous 5′P and 5′OH that are originally present in the untreated RNA samples. A cutting score can be calculated at each base which incorporates reads from P1 nuclease and reads from endogenous degradation or fragmentation products. A positive cutting score indicates that the base is single stranded.
Figure 5
Structural organization of the mRNA transcriptome. Thousands of yeast mRNAs are structurally probed in PARS and aligned according to their start and stop codons. The average PARS score of the coding sequence (CDS) is shown in blue; 5′ untranslated region (UTR) in yellow; 3′ UTR in red. The organization of secondary structures within the transcriptome revealed an increased accessibility of RNA structure near the start codon important for translation efficiency, shown by the negative spike. The coding sequence is more structured than the UTRs, as shown by the higher average (blue line), compared to the UTRs (orange and red lines). Some of these structures are important for cellular processes such as mRNA transport. A three nucleotide periodicity (inset box) in RNA is also seen in the coding region and is absent from the UTRs.
Similar articles
- Dawn of the in vivo RNA structurome and interactome.
Kwok CK. Kwok CK. Biochem Soc Trans. 2016 Oct 15;44(5):1395-1410. doi: 10.1042/BST20160075. Biochem Soc Trans. 2016. PMID: 27911722 Review. - StructureFold2: Bringing chemical probing data into the computational fold of RNA structural analysis.
Tack DC, Tang Y, Ritchey LE, Assmann SM, Bevilacqua PC. Tack DC, et al. Methods. 2018 Jul 1;143:12-15. doi: 10.1016/j.ymeth.2018.01.018. Epub 2018 Feb 2. Methods. 2018. PMID: 29410279 - Accurate detection of RNA stem-loops in structurome data reveals widespread association with protein binding sites.
Radecki P, Uppuluri R, Deshpande K, Aviran S. Radecki P, et al. RNA Biol. 2021 Oct 15;18(sup1):521-536. doi: 10.1080/15476286.2021.1971382. Epub 2021 Oct 4. RNA Biol. 2021. PMID: 34606413 Free PMC article. - Decoding the RNA structurome.
Lu Z, Chang HY. Lu Z, et al. Curr Opin Struct Biol. 2016 Feb;36:142-8. doi: 10.1016/j.sbi.2016.01.007. Epub 2016 Feb 26. Curr Opin Struct Biol. 2016. PMID: 26923056 Free PMC article. Review. - A combined sequence and structure based method for discovering enriched motifs in RNA from in vivo binding data.
Polishchuk M, Paz I, Kohen R, Mesika R, Yakhini Z, Mandel-Gutfreund Y. Polishchuk M, et al. Methods. 2017 Apr 15;118-119:73-81. doi: 10.1016/j.ymeth.2017.03.003. Epub 2017 Mar 6. Methods. 2017. PMID: 28274760
Cited by
- CentroidAlign-Web: A Fast and Accurate Multiple Aligner for Long Non-Coding RNAs.
Yonemoto H, Asai K, Hamada M. Yonemoto H, et al. Int J Mol Sci. 2013 Mar 18;14(3):6144-56. doi: 10.3390/ijms14036144. Int J Mol Sci. 2013. PMID: 23507751 Free PMC article. - The DEAD-box protein Dbp2 functions with the RNA-binding protein Yra1 to promote mRNP assembly.
Ma WK, Cloutier SC, Tran EJ. Ma WK, et al. J Mol Biol. 2013 Oct 23;425(20):3824-38. doi: 10.1016/j.jmb.2013.05.016. Epub 2013 May 28. J Mol Biol. 2013. PMID: 23721653 Free PMC article. - RNA-Seq Analysis Reveals Localization-Associated Alternative Splicing across 13 Cell Lines.
Zeng C, Hamada M. Zeng C, et al. Genes (Basel). 2020 Jul 18;11(7):820. doi: 10.3390/genes11070820. Genes (Basel). 2020. PMID: 32708427 Free PMC article. - Parallel computation of genome-scale RNA secondary structure to detect structural constraints on human genome.
Kawaguchi R, Kiryu H. Kawaguchi R, et al. BMC Bioinformatics. 2016 May 6;17(1):203. doi: 10.1186/s12859-016-1067-9. BMC Bioinformatics. 2016. PMID: 27153986 Free PMC article. - RASP: an atlas of transcriptome-wide RNA secondary structure probing data.
Li P, Zhou X, Xu K, Zhang QC. Li P, et al. Nucleic Acids Res. 2021 Jan 8;49(D1):D183-D191. doi: 10.1093/nar/gkaa880. Nucleic Acids Res. 2021. PMID: 33068412 Free PMC article.
References
- Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol. 2007;8:113–126. - PubMed
- Kozak M. Regulation of translation via mRNA structure in prokaryotes and eukaryotes. Gene. 2005;361:13–37. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous