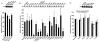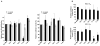Angiogenin-induced tRNA fragments inhibit translation initiation - PubMed (original) (raw)
Angiogenin-induced tRNA fragments inhibit translation initiation
Pavel Ivanov et al. Mol Cell. 2011.
Abstract
Angiogenin is a stress-activated ribonuclease that cleaves tRNA within anticodon loops to produce tRNA-derived stress-induced fragments (tiRNAs). Transfection of natural or synthetic tiRNAs inhibits protein synthesis and triggers the phospho-eIF2α-independent assembly of stress granules (SGs), essential components of the stress response program. We show that selected tiRNAs inhibit protein synthesis by displacing eIF4G/eIF4A from uncapped > capped RNAs. tiRNAs also displace eIF4F, but not eIF4E:4EBP1, from isolated m(7)G cap. We identify a terminal oligoguanine motif that is required to displace the eIF4F complex, inhibit translation, and induce SG assembly. We show that the tiRNA-associated translational silencer YB-1 contributes to angiogenin-, tiRNA-, and oxidative stress-induced translational repression. Our data reveal some of the mechanisms by which stress-induced tRNA cleavage inhibits protein synthesis and activates a cytoprotective stress response program.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures
Figure 1
5′-tiRNAs inhibit translation of mRNA reporters in vitro. A. Natural 5′-tiRNAs. Uncapped Firefly luciferase mRNA (Promega) was translated in RRL in the presence of control RNA mix (Ctrl-1-2-3; derived from piRNAs or random sequences), natural 5′-tiRNAs (Nat 5′end) or natural 3′-tiRNAs (Nat 3′end). Luciferase expression is relative to that in the absence of RNA (No RNA =100%). Means and standard deviations are from three independent experiments (*-p = 0.01–0.02 (Table S1), Student’s t-test, n=3). NB: Northern Blotting for the luciferase mRNA reporter. For P-values, see Table S1. B. Synthetic tiRNAs. Uncapped Firefly luciferase expression in the presence of synthetic 5′- and 3′-tiRNAs (100 picomoles/10 μl) as described in Fig. 1A. Means and standard deviations are from four independent experiments (*- p < 0.05 (Table SI), Student’s t-test, n=4). These reactions contain 50 ng/10 μl of uncapped Firefly RNA. NB: Northern Blotting for the luciferase mRNA reporter. See also Figure S1. C. Synthetic 5′-tiRNAAla inhibits translation of capped Firefly luciferase mRNA. Capped Firefly luciferase RNA (10 ng/10 μl) was translated in RRL in the presence of indicated RNAs (100 picomoles/10 μl) in the absence (dark bars) or presence (light bars) of cap analogue (m7GpppG, 0.1mM). Means and standard deviations are from three independent experiments (*-p < 0.05 (Table SI); # p=0.04, comparing 5′-tiRNAAla in the absence or presence of the cap analogue, Student’s t-test, n=3). Cap analogue reduces the basal level of capped mRNA translation by ~2.5 fold. NB: Northern Blotting for the capped luciferase mRNA reporter.
Figure 2
Effect of tiRNAs on EMCV IRES-mediated translation initiaton. A. Synthetic 5′-tiRNAAla does not inhibit EMCV IRES-driven translation. Uncapped (dark bars) or capped (light bars) pF/R bicistronic transcripts were translated in RRL. Left panel: The relative ratio of Firefly to Renilla counts. Means and standard deviations are from 3–4 independent experiments (*-p <0.05, compared to no RNA and three control RNAs (ctrl-1,-2,-3) (Tables S1 and S2, Student’s t-test, n=3–4). Right panel: counts derived from EMCV IRES-driven translation of Renilla ORF relative to the no RNA control (100%). For actual luciferase counts, see Table S2. B. Synthetic 5′-tiRNAAla inhibits translation of EMCV IRES UA7 variant. Uncapped monocistronic transcripts encoding Renilla luciferase under control of different EMCV IRES variants (WT EMCV-UA6 (upper panel) and EMCV-UA7 (lower panel)) were translated in RRL in the presence of control RNAs (ctrl-1,-2,-3), 5′-tiRNAAla or 3′-tiRNAAla. Means and standard deviations are from three independent experiments *-p <0.05, compared to no RNA and three control RNAs (ctrl-1,-2,-3) (Table SI, Student’s t-test, n=3). See also Figure S2. For actual luciferase counts, see Table S2.
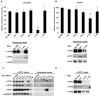
Figure 3
5′-tiRNAs target eIF4F complex. A. Synthetic 5′-tiRNAAla displaces eIF4G from uncapped mRNA. Upper panel: Uncapped polyA-biotinylated transcript encoding Renilla luciferase was translated in RRL supplemented with U2OS extract in the presence of the indicated control RNAs or tiRNAs. Luciferase expression is relative to that in the absence of RNA (No RNA =100%). Means and standard deviations are from three independent experiments. *-p <0.05, compared to no RNA and three control RNAs (ctrl-1,-2,-3) (Table S1, Student’s t-test, n=3). Lower panel: Streptavidin pull-down of uncapped polyA-biotinylated transcript. In vitro translation was performed as indicated in the upper panel in the presence of control RNA (Ctrl), 5′-tiRNAAla or cap analogue (Cap, m7GpppG, 0.1mM). Streptavidin beads were used to pull down reporter RNA-protein complexes. Bound proteins were identified by western blotting using indicated antibodies. For p values, see Table S1. B. Synthetic 5′-tiRNAAla displaces eIF4G from capped mRNA. Upper panel: Capped polyA-biotinylated transcript encoding Renilla luciferase was translated in RRL supplemented with U2OS extract in the presence of the indicated control RNAs or tiRNAs. Luciferase expression is relative to that in the absence of RNA (No RNA =100%). Means and standard deviations are from three independent experiments. *-p <0.05, compared to no RNA and three control RNAs (ctrl-1,-2,-3) (Table S1, Student’s t-test, n=3). Lower panel: Streptavidin pull-down of capped polyA-biotinylated transcript as in Fig. 3A. C. 5′-tiRNAAla displaces the eIF4F complex from m7GTP-Sepharose. The indicated 3′-end biotinylated RNAs or cap analogue (Cap, m7GpppG, 0.1mM) were added to m7GTP-sepharose complexes and displaced components were quantified by western blot (left panel). Streptavidin beads were used to pull down displaced RNA-bound proteins and were identified by western blotting (right panel). D. eIF4F complex was assembled on m7GTP-Sepharose from RRL supplemented with U2OS extract. No RNA, control RNA (ctrl), 5′-tiRNAAla or 5′-tiRNACys were added to m7GTP-sepharose complexes. Integrity of eIF4F complex was monitored by western blotting using anti-eIF4E, -eIF4G and –eIF4A antibodies.
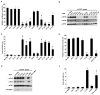
Figure 4
Structure/function analysis of tiRNAs. A. In vitro translation in RRL. Uncapped Firefly luciferase mRNA was translated in RRL in the presence of synthetic control RNAs (ctrl-1,-2,-3), 5′-tiRNAAla, or the indicated mutants. Luciferase expression is relative to that produced in the absence of any RNA (No RNA = 100%). Means and standard deviations are from four independent experiments (*- p < 0.05 (Table SI), Student’s t-test, n=4). See also Figures S3 and S4. For p values, see Table S1. B. Displacement of eIF4F complexes from m7GTP-sepharose. eIF4F complex was assembled on m7GTP-sepharose in U2OS lysates. The indicated synthetic RNAs were added to beads and analyzed as described in Figure 3C. C. SG assembly. U2OS cells were transfected with indicated RNAs. The percentage of U2OS cells with SGs was quantified by counting 200 cells/experiment. Error bars reflect standard deviations of the mean (*-p-value < 0.05 (Table S1), relative to control without any RNA, Student’s t-test, n=3). D. Addition of 5′-TOG motif “activates” 5′-tiRNAMet to inhibit translation in RRL as in Figure 3C. Error bars reflect standard deviations of the mean (*-p-value < 0.05 (Table S1), relative to control without any RNA, Student’s t-test, n=3). E. 5′-TOG-containing tiRNAs displace eIF4F. Western blot analysis was used to quantify the binding of eIF4F complexes and eIF4E-BP1 as in Figure 4B. F. Addition of 5′-TOG “activates” 5′-tiRNAMet to induce the assembly of SGs as in Figure 3C. Error bars reflect standard deviations of the mean (*-p-value < 0.05 (Table S1), relative to control without any RNA, Student’s t-test, n=3).
Figure 5
Identification of tiRNA-interacting proteins. A. Validation of selected candidate 5′-tiRNAAla-binding proteins. Proteins bound to biotinylated control RNA and 5′-tiRNAAla were purified using streptavidine beads, subjected to the indicated salt washes (0.1M, 0.3M and 0.5M), eluted, then analyzed by Western blotting using corresponding antibodies. For list of interacting proteins, see Table S3. B. tiRNA-binding proteins required for 5′-tiRNAAla-induced SG assembly. U2OS cells treated with indicated siRNAs, were mock transfected (No RNA) or transfected with control RNA (Ctrl) or 5′-tiRNAAla (5′-Ala) before processing for immunofluorescence microscopy to quantify SGs. Left panel: Efficiency of knock down was assessed by comparing the expression of each protein from siRNA control transfections (Ctrl, serial dilutions) to targeted siRNA transfections (Target). Tubulin was used as a specificity and loading control. Right panel: Percentage of cells with SGs was quantified by counting 200 cells/experiment. Results show the means and standard deviations from the indicated number of experiments (AGO2 siRNA, n=7; PABP1 siRNA, n=3; YB-1 siRNA, n=9; Vigilin siRNA, n=3; FXR1 siRNA, n=3). *-p-value = 0.0001 when comparing the percentage of cells with SGs in control and YB-1 siRNA treated cells; no other treatment reached statistical significance. C. YB-1 is a part of complex displaced by 5′-tiRNAAla from m7GTP-sepharose. Upper panel: Biotinylated control RNA (Ctrl-bio), 5′-tiRNAAla (5′-Ala-bio) or its mutants (U4G-bio and UU3G-bio) were immobilized on streptavidin beads and used to pull down protein complexes as described in Fig. 4A. Bound complexes were analyzed by Western blotting using anti-YB-1 antibodies. Lower panel: eIF4E-containing complexes were assembled on m7GTP-Sepharose and incubated with no RNA, biotinylated control RNA (Ctrl) or biotinylated 5′-tiRNAAla (5′Ala). Beads were washed, bound proteins were eluted and analyzed by Western blot using antibodies against eIF4E and YB-1 (left panel). Proteins associated with biotin-RNAs were pulled down using streptavidin beads, eIF4E and YB-1 were detected by western blotting (right panel). D. YB-1 is required for 5′-tiRNA-induced translational repression. U2OS cells treated with control or YB-1 siRNAs were transfected with Ctrl RNA, 5′-tiRNAAla, 5′-tiRNACys, 5′-tiRNAMet, and TOG-5′-Met and then labeled with (35S)-methionine. (35S)-methionine incorporation in cells transfected with no RNA is reported as 100%. Error bars show means and standard deviations (5′-tiRNAAla, p=0.002, n=10; 5′-tiRNACys, p=0.01, n=4; when comparing inhibition of global protein synthesis by tiRNAs in control and YB-1 siRNA treated cells; no other treatments reached statistical significance). E. YB-1 is required for stress- and angiogenin-induced translational repression. U2OS treated with control or YB-1 siRNAs were labeled with (35S)-methionine in the presence of sodium arsenite (SA), Pateamine A (PatA), hydrogen peroxide (H2O2), angiogenin (ANG), heat inactivated ANG (HI ANG), angiogenin-neutralizing antibody (26-2F) alone or in combination with ANG (ANG+26-2F). (35S)-methionine incorporation in untreated cells (No treat) is reported as 100%. Error bars show means and standard deviations (SA, p=0.0003, n=8; ANG, p=0.002, n=8; and PatA, p=0.04, n=3 when comparing inhibition of global protein synthesis by those treatments in control and YB-1 siRNA treated cells; *-p-value = 0.003; n=4 and 0.01; n=3 when comparing inhibition of global protein synthesis by ANG+26-2F and HI ANG to angiogenin-treated cells, respectively).
Comment in
- Functional expansion of the tRNA world under stress.
Yang XL, Schimmel P. Yang XL, et al. Mol Cell. 2011 Aug 19;43(4):500-2. doi: 10.1016/j.molcel.2011.08.004. Mol Cell. 2011. PMID: 21855789 Free PMC article.
Similar articles
- G-quadruplex structures contribute to the neuroprotective effects of angiogenin-induced tRNA fragments.
Ivanov P, O'Day E, Emara MM, Wagner G, Lieberman J, Anderson P. Ivanov P, et al. Proc Natl Acad Sci U S A. 2014 Dec 23;111(51):18201-6. doi: 10.1073/pnas.1407361111. Epub 2014 Nov 17. Proc Natl Acad Sci U S A. 2014. PMID: 25404306 Free PMC article. - Angiogenin cleaves tRNA and promotes stress-induced translational repression.
Yamasaki S, Ivanov P, Hu GF, Anderson P. Yamasaki S, et al. J Cell Biol. 2009 Apr 6;185(1):35-42. doi: 10.1083/jcb.200811106. Epub 2009 Mar 30. J Cell Biol. 2009. PMID: 19332886 Free PMC article. - YB-1 regulates tiRNA-induced Stress Granule formation but not translational repression.
Lyons SM, Achorn C, Kedersha NL, Anderson PJ, Ivanov P. Lyons SM, et al. Nucleic Acids Res. 2016 Aug 19;44(14):6949-60. doi: 10.1093/nar/gkw418. Epub 2016 May 12. Nucleic Acids Res. 2016. PMID: 27174937 Free PMC article. - The Many Virtues of tRNA-derived Stress-induced RNAs (tiRNAs): Discovering Novel Mechanisms of Stress Response and Effect on Human Health.
Saikia M, Hatzoglou M. Saikia M, et al. J Biol Chem. 2015 Dec 11;290(50):29761-8. doi: 10.1074/jbc.R115.694661. Epub 2015 Oct 13. J Biol Chem. 2015. PMID: 26463210 Free PMC article. Review. - tRNA fragmentation and protein translation dynamics in the course of kidney injury.
Mami I, Pallet N. Mami I, et al. RNA Biol. 2018;15(9):1147-1156. doi: 10.1080/15476286.2015.1107704. Epub 2018 Feb 12. RNA Biol. 2018. PMID: 26513712 Free PMC article. Review.
Cited by
- tRNA-derived fragments as novel potential biomarkers for relapsed/refractory multiple myeloma.
Xu C, Liang T, Zhang F, Liu J, Fu Y. Xu C, et al. BMC Bioinformatics. 2021 May 11;22(1):238. doi: 10.1186/s12859-021-04167-8. BMC Bioinformatics. 2021. PMID: 33971811 Free PMC article. - 5' tRNA halves are present as abundant complexes in serum, concentrated in blood cells, and modulated by aging and calorie restriction.
Dhahbi JM, Spindler SR, Atamna H, Yamakawa A, Boffelli D, Mote P, Martin DI. Dhahbi JM, et al. BMC Genomics. 2013 May 2;14:298. doi: 10.1186/1471-2164-14-298. BMC Genomics. 2013. PMID: 23638709 Free PMC article. - Assessment of small RNA sorting into different extracellular fractions revealed by high-throughput sequencing of breast cell lines.
Tosar JP, Gámbaro F, Sanguinetti J, Bonilla B, Witwer KW, Cayota A. Tosar JP, et al. Nucleic Acids Res. 2015 Jun 23;43(11):5601-16. doi: 10.1093/nar/gkv432. Epub 2015 May 4. Nucleic Acids Res. 2015. PMID: 25940616 Free PMC article. - Downregulation of angiogenin inhibits the growth and induces apoptosis in human bladder cancer cells through regulating AKT/mTOR signaling pathway.
Shu J, Huang M, Tian Q, Shui Q, Zhou Y, Chen J. Shu J, et al. J Mol Histol. 2015 Apr;46(2):157-71. doi: 10.1007/s10735-014-9608-x. Epub 2015 Jan 7. J Mol Histol. 2015. PMID: 25564356 - Endogenous tRNA-Derived Fragments Suppress Breast Cancer Progression via YBX1 Displacement.
Goodarzi H, Liu X, Nguyen HC, Zhang S, Fish L, Tavazoie SF. Goodarzi H, et al. Cell. 2015 May 7;161(4):790-802. doi: 10.1016/j.cell.2015.02.053. Cell. 2015. PMID: 25957686 Free PMC article.
References
- Bochkov YA, Palmenberg AC. Translational efficiency of EMCV IRES in bicistronic vectors is dependent upon IRES sequence and gene location. BioTechniques. 2006;41:283–284. 286, 288. passim. - PubMed
- Evdokimova V, Ovchinnikov LP, Sorensen PH. Y-box binding protein 1: providing a new angle on translational regulation. Cell cycle (Georgetown, Tex. 2006;5:1143–1147. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R56 AI033600/AI/NIAID NIH HHS/United States
- AI065858/AI/NIAID NIH HHS/United States
- P01 AI065858/AI/NIAID NIH HHS/United States
- R01 AI033600/AI/NIAID NIH HHS/United States
- AI033600/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous