Recombination in IS26 and Tn2 in the evolution of multiresistance regions carrying blaCTX-M-15 on conjugative IncF plasmids from Escherichia coli - PubMed (original) (raw)
Recombination in IS26 and Tn2 in the evolution of multiresistance regions carrying blaCTX-M-15 on conjugative IncF plasmids from Escherichia coli
Sally R Partridge et al. Antimicrob Agents Chemother. 2011 Nov.
Abstract
CTX-M-15 now appears to be the dominant extended-spectrum β-lactamase worldwide, and a number of different factors may contribute to this success. These include associations between bla(CTX-M-15) and particular plasmids (IncF) and/or strains, such as Escherichia coli ST131, as well as the genetic contexts in which this gene is found. We previously identified bla(CTX-M-15) as the dominant ESBL gene in the western Sydney area, Australia, and found that it was carried mainly on IncF or IncI1 plasmids. Here, we have mapped the multiresistance regions of the 11 conjugative plasmids with one or more IncF replicons obtained from that survey and conducted a limited comparison of plasmid backbones. Two plasmids with only an IncFII replicon appear to be very similar to the published plasmids pC15-1a and pEK516. The remaining nine plasmids, with multiple IncF replicons, have multiresistance regions related to those of pC15-1a and pEK516, but eight contain additional modules previously found in resistance plasmids from different geographic locations that carry a variety of different resistance genes. Differences between the multiresistance regions are largely due to IS26-mediated deletions, insertions, and/or rearrangements, which can explain the observed variable associations between bla(CTX-M-15) and certain other resistance genes. We found no evidence of independent movement of bla(CTX-M-15) or of a large multiresistance region between different plasmid backbones. Instead, homologous recombination between common components, such as IS26 and Tn2, appeared to be more important in creating new multiresistance regions, and this may be coupled with recombination in plasmid backbones to reassort multiple IncF replicons as well as components of multiresistance regions.
Figures
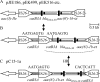
Fig. 1.
MRRs carrying bla_CTX-M-15. Similar structures are grouped together. Different transposons and other modules have different shading and are generally labeled only once (tnp21 and mer21, transposition and mercury resistance regions of Tn_21, respectively). ISs are labeled with their number/name, with the pointed end indicating IRR. Tall bars represent the 38-bp IR of transposons, as indicated. Positions/orientations of selected resistance and other genes are indicated by arrows, generally labeled only once. Abbreviations: A, tnpA; R, tnpR; 1b, bla_TEM-1b, 1c, bla_TEM-1c; 15, bla_CTX-M-15; IIe, aac(3)-IIe; cr, aac(6_′)-Ib -cr; 30, bla_OXA-30. Class 1 integron components are indicated as follows: 5′, 5′-CS; 3′, 3′-CS; narrow boxes, gene cassettes; small black boxes, attC sites; 1, dfrA17-aadA5 cassette array; 2, dfrA12-gcuF-aadA2 cassette array; tni402, transposition region of Tn_402; IRi, 25-bp IR at intI1 end; IRt, 25-bp IR at tni end. The chrA_-mph(A) module (indicated in panel D) is found after position 1593 of the 3′-CS and consists of part of a chromate resistance transposon (IR_chr and chrA), 123 bp of the IRt end of tni402, IS_6100, and the mph(A)-mrx-mphR(A) macrolide resistance region (29). Dashed lines represent the IncFII backbone. The nqrC_-like (putative Na+-translocating NADH-quinone reductase) and scsD (secreted copper sensitivity repressor) genes are part of an ∼10-kb region found in several IncF plasmids that is apparently derived from the Citrobacter koseri chromosome (GenBank accession no. CP000822). Arrows labeled HR and dotted lines indicate where homologous recombination explains differences between structures. (A) pC15-1a MRR (AY458016). DRs flanking the IS_Ecp1_-bla_CTX-M-15 transposition unit are shown. The left-hand boundary with the IncFII backbone is defined by a remnant of IS_1. (B) Related MRRs in pJIE186, pEK516 (EU935738; rearranged version; see Fig. S3B in the supplemental material), and pJIE100. (C) pJIE098 MRR. (D) pJIE118/pJIE157/pJIE224 MRR. The IRi-tnp21 boundary is the same as in Tn_21 (AF071413). (E) pJIE286 and pEK499 MRRs. The separate region shown on the right is in pEK499 (EU935739) only. (F) Related MRRs in pJIE085, pJIE134, and pJIE250. IS_26 at the end of chrA-mph(A) truncates a region derived from IncQ plasmids that includes sul2 and a truncated Tn_5393_, carrying the strAB genes, and ends with IR_str_ (48). This truncates Tn_1721_, carrying tetAR(A), which is followed by part of tni402 and a region flanked by two copies of IS_26_ apparently derived from a larger region found in pU302L (AY333434) and pCTX-M3 (AF550415), in which IS_Cfr1_ and orf69 are complete. The pJIE085 and pJIE134 MRRs end with a complex multi-IS structure (details in Fig. S5A in the supplemental material), while pJIE250 has IS_26_ only, truncating the cjrA gene found in several plasmids. (G) pJIE101 MRR. resP is identical to the genes in IncN plasmids. IS_26_ at the end of chrA-mph(A) forms part of composite transposon Tn_4352_ (46), the IS_26_-tni402 boundary is in >30 sequences in GenBank, the IRt_-mer21_ boundary is the same as in Tn_21_, and the _mer21_-IRTEM boundary is seen in other plasmid sequences (25, 30).
Fig. 2.
IS_26_-mediated rearrangement of the cassette array containing aac(6_′)-Ib_ -cr, _bla_OXA-30, and catB3_Δ. Components are generally represented as described in the legend to Fig. 1. (A) Cassette array region in MRRs of pJIE186 and eight other pJIE plasmids, pEK499, pEK516, pEC_L8, and pEC_L46; (B) insertion of IS_26_-3 into the catB3 Δ cassette would generate 8-bp DRs characteristic of IS_26 transposition (9); (C) subsequent homologous recombination between IS_26_-2 and IS_26_-3 (dotted line) would invert the intervening region to give the pC15-1a configuration, in which the 8-bp sequence adjacent to IRL of IS_26_-3 is the reverse complement of the 8 bp adjacent to IRR of IS_26_-2.
Fig. 3.
Deletions in MRRs caused by insertion of additional copies of IS_26_ and homologous recombination. Components are generally represented as described in the legend to Fig. 1. (A) Part of the pJIE186 MRR; (B) insertion of an extra copy of IS_26_ in Tn_5403_ and homologous recombination with the IS_26_ element interrupting the IR_tnp_ end of Tn_2_ (dotted line) would result in the pJIE100 structure; (C) insertion of an extra copy of IS_26_ in Tn_1721_ and homologous recombination with the IS_26_ element next to aac(3)-IIe would result in the pJIE098 structure.
Fig. 4.
Recombination between common MRR modules. Components are generally represented as described in the legend to Fig. 1. Crosses indicate regions where homologous recombination could have taken place. (A) Relationship between the pJIE085 and pKF3-140 MRRs. An extra 198 bp of the 5′-CS is present in pKF3-140 compared with pJIE085, as indicated. The IS_26_ element truncating orf69 and Tn_2_ in pJIE085 marks the end of the match to the pKF3-140 MRR. (B) Relationship between the pJIE101 MRR, TnSF1, and IncN plasmids. The vertical arrow in TnSF1 indicates a 521-bp insert not present in any other examples of chrA-mph(A). The cassette arrays in pJIE101, TnSF1, and IncN plasmids are indicated. The pJIE101 IncN replicon is 100% identical to p12 (14) over the region amplified. Sequences referred to in this diagram are from the following GenBank accession numbers: pKF3-140, FJ876827; TnSF1, AF188331; pKP96, EU195449; p 12, FJ223605; pKOX105, HM126016; and pK245, DQ449578.
Similar articles
- Different IncI1 plasmids from Escherichia coli carry ISEcp1-blaCTX-M-15 associated with different Tn2-derived elements.
Zong Z, Ginn AN, Dobiasova H, Iredell JR, Partridge SR. Zong Z, et al. Plasmid. 2015 Jul;80:118-26. doi: 10.1016/j.plasmid.2015.04.007. Epub 2015 Apr 27. Plasmid. 2015. PMID: 25929173 - Advantage of the F2:A1:B- IncF Pandemic Plasmid over IncC Plasmids in In Vitro Acquisition and Evolution of _bla_CTX-M Gene-Bearing Plasmids in Escherichia coli.
Mahérault AC, Kemble H, Magnan M, Gachet B, Roche D, Le Nagard H, Tenaillon O, Denamur E, Branger C, Landraud L. Mahérault AC, et al. Antimicrob Agents Chemother. 2019 Sep 23;63(10):e01130-19. doi: 10.1128/AAC.01130-19. Print 2019 Oct. Antimicrob Agents Chemother. 2019. PMID: 31332067 Free PMC article. - IncI1 plasmids carrying bla(CTX-M-1) or bla(CMY-2) genes in Escherichia coli from healthy humans and animals in Tunisia.
Ben Sallem R, Ben Slama K, Rojo-Bezares B, Porres-Osante N, Jouini A, Klibi N, Boudabous A, Sáenz Y, Torres C. Ben Sallem R, et al. Microb Drug Resist. 2014 Oct;20(5):495-500. doi: 10.1089/mdr.2013.0224. Epub 2014 May 14. Microb Drug Resist. 2014. PMID: 24826863 - Antibiotic resistance in Escherichia coli in husbandry animals: the African perspective.
Alonso CA, Zarazaga M, Ben Sallem R, Jouini A, Ben Slama K, Torres C. Alonso CA, et al. Lett Appl Microbiol. 2017 May;64(5):318-334. doi: 10.1111/lam.12724. Lett Appl Microbiol. 2017. PMID: 28208218 Review. - The Significance of Epidemic Plasmids in the Success of Multidrug-Resistant Drug Pandemic Extraintestinal Pathogenic Escherichia coli.
Pitout JDD, Chen L. Pitout JDD, et al. Infect Dis Ther. 2023 Apr;12(4):1029-1041. doi: 10.1007/s40121-023-00791-4. Epub 2023 Mar 22. Infect Dis Ther. 2023. PMID: 36947392 Free PMC article. Review.
Cited by
- Evolutionary History of the Global Emergence of the Escherichia coli Epidemic Clone ST131.
Stoesser N, Sheppard AE, Pankhurst L, De Maio N, Moore CE, Sebra R, Turner P, Anson LW, Kasarskis A, Batty EM, Kos V, Wilson DJ, Phetsouvanh R, Wyllie D, Sokurenko E, Manges AR, Johnson TJ, Price LB, Peto TE, Johnson JR, Didelot X, Walker AS, Crook DW; Modernizing Medical Microbiology Informatics Group (MMMIG). Stoesser N, et al. mBio. 2016 Mar 22;7(2):e02162. doi: 10.1128/mBio.02162-15. mBio. 2016. PMID: 27006459 Free PMC article. - Characterization of blaCTX-M sequences of Indian origin and thirteen uropathogenic Escherichia coli isolates resistant to multiple antibiotics.
Siddaramappa S, Pullela K, Thimmappa B, Devkota R, Bajaj R, Manivannan B, Mahalingam N, Pradeep BE. Siddaramappa S, et al. BMC Res Notes. 2018 Aug 31;11(1):630. doi: 10.1186/s13104-018-3735-5. BMC Res Notes. 2018. PMID: 30170618 Free PMC article. - Prevalence and characteristics of the epidemic multiresistant Escherichia coli ST131 clonal group among extended-spectrum beta-lactamase-producing E. coli isolates in Copenhagen, Denmark.
Olesen B, Hansen DS, Nilsson F, Frimodt-Møller J, Leihof RF, Struve C, Scheutz F, Johnston B, Krogfelt KA, Johnson JR. Olesen B, et al. J Clin Microbiol. 2013 Jun;51(6):1779-85. doi: 10.1128/JCM.00346-13. Epub 2013 Apr 3. J Clin Microbiol. 2013. PMID: 23554186 Free PMC article. - Extended-spectrum β-lactamase-encoding genes are spreading on a wide range of Escherichia coli plasmids existing prior to the use of third-generation cephalosporins.
Branger C, Ledda A, Billard-Pomares T, Doublet B, Fouteau S, Barbe V, Roche D, Cruveiller S, Médigue C, Castellanos M, Decré D, Drieux-Rouze L, Clermont O, Glodt J, Tenaillon O, Cloeckaert A, Arlet G, Denamur E. Branger C, et al. Microb Genom. 2018 Sep;4(9):e000203. doi: 10.1099/mgen.0.000203. Epub 2018 Aug 6. Microb Genom. 2018. PMID: 30080134 Free PMC article. - Epidemiology of Plasmid Lineages Mediating the Spread of Extended-Spectrum Beta-Lactamases among Clinical Escherichia coli.
Mahmud B, Wallace MA, Reske KA, Alvarado K, Muenks CE, Rasmussen DA, Burnham CD, Lanzas C, Dubberke ER, Dantas G. Mahmud B, et al. mSystems. 2022 Oct 26;7(5):e0051922. doi: 10.1128/msystems.00519-22. Epub 2022 Aug 22. mSystems. 2022. PMID: 35993734 Free PMC article.
References
- Bailey J. K., Pinyon J. L., Anantham S., Hall R. M. 2011. Distribution of the blaTEM gene and blaTEM-containing transposons in commensal Escherichia coli. J. Antimicrob. Chemother. 66:745–751 - PubMed
- Baquero F. 2004. From pieces to patterns: evolutionary engineering in bacterial pathogens. Nat. Rev. Microbiol. 2:510–518 - PubMed
- Barton B. M., Harding G. P., Zuccarelli A. J. 1995. A general method for detecting and sizing large plasmids. Anal. Biochem. 226:235–240 - PubMed
- Baudry P. J., et al. 2009. Mechanisms of resistance and mobility among multidrug-resistant CTX-M-producing Escherichia coli from Canadian intensive care units: the 1st report of QepA in North America. Diagn. Microbiol. Infect. Dis. 63:319–326 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases