Population genetics of Vibrio cholerae from Nepal in 2010: evidence on the origin of the Haitian outbreak - PubMed (original) (raw)
. 2011 Sep 1;2(4):e00157-11.
doi: 10.1128/mBio.00157-11. Print 2011.
Lance B Price, James M Schupp, John D Gillece, Rolf S Kaas, David M Engelthaler, Valeria Bortolaia, Talima Pearson, Andrew E Waters, Bishnu Prasad Upadhyay, Sirjana Devi Shrestha, Shailaja Adhikari, Geeta Shakya, Paul S Keim, Frank M Aarestrup
Affiliations
- PMID: 21862630
- PMCID: PMC3163938
- DOI: 10.1128/mBio.00157-11
Population genetics of Vibrio cholerae from Nepal in 2010: evidence on the origin of the Haitian outbreak
Rene S Hendriksen et al. mBio. 2011.
Abstract
Cholera continues to be an important cause of human infections, and outbreaks are often observed after natural disasters, such as the one following the 2010 earthquake in Haiti. Once the cholera outbreak was confirmed, rumors spread that the disease was brought to Haiti by a battalion of Nepalese soldiers serving as United Nations peacekeepers. This possible connection has never been confirmed. We used whole-genome sequence typing (WGST), pulsed-field gel electrophoresis (PFGE), and antimicrobial susceptibility testing to characterize 24 recent Vibrio cholerae isolates from Nepal and evaluate the suggested epidemiological link with the Haitian outbreak. The isolates were obtained from 30 July to 1 November 2010 from five different districts in Nepal. We compared the 24 genomes to 10 previously sequenced V. cholerae isolates, including 3 from the Haitian outbreak (began July 2010). Antimicrobial susceptibility and PFGE patterns were consistent with an epidemiological link between the isolates from Nepal and Haiti. WGST showed that all 24 V. cholerae isolates from Nepal belonged to a single monophyletic group that also contained isolates from Bangladesh and Haiti. The Nepalese isolates were divided into four closely related clusters. One cluster contained three Nepalese isolates and three Haitian isolates that were almost identical, with only 1- or 2-bp differences. Results in this study are consistent with Nepal as the origin of the Haitian outbreak. This highlights how rapidly infectious diseases might be transmitted globally through international travel and how public health officials need advanced molecular tools along with standard epidemiological analyses to quickly determine the sources of outbreaks.
Figures
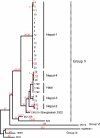
FIG 1
Genetic relationships among V. cholerae isolates from Nepal and Haiti. A single maximum parsimony tree was reconstructed using 752 SNPs from 34 whole-genome sequences. There were 184 parsimony-informative SNPs, of which 6 were homoplastic, resulting in a CI of 0.97 (excluding uninformative characters). The branch lengths are labeled in red, and for branches affected by homoplasy, minimum and maximum branch lengths are designated. Members of SNP genotypic group V (16) are indicated. SNP differences among the three most closely related Nepali groups and the Haitian group are shown and characterized in Table S1 in the supplemental material.
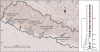
FIG 2
Locations of the five districts in Nepal where the V. cholerae O1 Ogawa strains were isolated.
Comment in
- South Asia instead of Nepal may be the origin of the Haitian cholera outbreak strains.
Pun SB. Pun SB. mBio. 2011 Oct 11;2(5):e00219-11. doi: 10.1128/mBio.00219-11. Print 2011. mBio. 2011. PMID: 21990616 Free PMC article. No abstract available.
Similar articles
- Deciphering the origins and tracking the evolution of cholera epidemics with whole-genome-based molecular epidemiology.
Grad YH, Waldor MK. Grad YH, et al. mBio. 2013 Sep 10;4(5):e00670-13. doi: 10.1128/mBio.00670-13. mBio. 2013. PMID: 24023387 Free PMC article. - Genomic epidemiology of the Haitian cholera outbreak: a single introduction followed by rapid, extensive, and continued spread characterized the onset of the epidemic.
Eppinger M, Pearson T, Koenig SS, Pearson O, Hicks N, Agrawal S, Sanjar F, Galens K, Daugherty S, Crabtree J, Hendriksen RS, Price LB, Upadhyay BP, Shakya G, Fraser CM, Ravel J, Keim PS. Eppinger M, et al. mBio. 2014 Nov 4;5(6):e01721. doi: 10.1128/mBio.01721-14. mBio. 2014. PMID: 25370488 Free PMC article. - The origin of the Haitian cholera outbreak strain.
Chin CS, Sorenson J, Harris JB, Robins WP, Charles RC, Jean-Charles RR, Bullard J, Webster DR, Kasarskis A, Peluso P, Paxinos EE, Yamaichi Y, Calderwood SB, Mekalanos JJ, Schadt EE, Waldor MK. Chin CS, et al. N Engl J Med. 2011 Jan 6;364(1):33-42. doi: 10.1056/NEJMoa1012928. Epub 2010 Dec 9. N Engl J Med. 2011. PMID: 21142692 Free PMC article. - Widespread epidemic cholera caused by a restricted subset of Vibrio cholerae clones.
Moore S, Thomson N, Mutreja A, Piarroux R. Moore S, et al. Clin Microbiol Infect. 2014 May;20(5):373-9. doi: 10.1111/1469-0691.12610. Epub 2014 Mar 29. Clin Microbiol Infect. 2014. PMID: 24575898 Review. - Circulation and transmission of clones of Vibrio cholerae during cholera outbreaks.
Stine OC, Morris JG Jr. Stine OC, et al. Curr Top Microbiol Immunol. 2014;379:181-93. doi: 10.1007/82_2013_360. Curr Top Microbiol Immunol. 2014. PMID: 24407776 Free PMC article. Review.
Cited by
- Rapid and robust phylotyping of spa t003, a dominant MRSA clone in Luxembourg and other European countries.
Engelthaler DM, Kelley E, Driebe EM, Bowers J, Eberhard CF, Trujillo J, Decruyenaere F, Schupp JM, Mossong J, Keim P, Even J. Engelthaler DM, et al. BMC Infect Dis. 2013 Jul 23;13:339. doi: 10.1186/1471-2334-13-339. BMC Infect Dis. 2013. PMID: 23879266 Free PMC article. - Non-O1 Vibrio cholerae unlinked to cholera in Haiti.
Mekalanos JJ, Robins W, Ussery DW, Davis BM, Schadt E, Waldor MK. Mekalanos JJ, et al. Proc Natl Acad Sci U S A. 2012 Nov 20;109(47):E3206; author reply E3207. doi: 10.1073/pnas.1212443109. Epub 2012 Oct 3. Proc Natl Acad Sci U S A. 2012. PMID: 23035253 Free PMC article. No abstract available. - Bacterial genomes in epidemiology--present and future.
Croucher NJ, Harris SR, Grad YH, Hanage WP. Croucher NJ, et al. Philos Trans R Soc Lond B Biol Sci. 2013 Feb 4;368(1614):20120202. doi: 10.1098/rstb.2012.0202. Print 2013 Mar 19. Philos Trans R Soc Lond B Biol Sci. 2013. PMID: 23382424 Free PMC article. Review. - Geographical information software and shopper card data, aided in the discovery of a Salmonella Enteritidis outbreak associated with Turkish pine nuts.
Bedard B, Kennedy BS, Weimer AC. Bedard B, et al. Epidemiol Infect. 2014 Dec;142(12):2567-71. doi: 10.1017/S0950268814000223. Epub 2014 Feb 14. Epidemiol Infect. 2014. PMID: 24534462 Free PMC article. - Metaphylogenetic analysis of global sewage reveals that bacterial strains associated with human disease show less degree of geographic clustering.
Ahrenfeldt J, Waisi M, Loft IC, Clausen PTLC, Allesøe R, Szarvas J, Hendriksen RS, Aarestrup FM, Lund O. Ahrenfeldt J, et al. Sci Rep. 2020 Feb 20;10(1):3033. doi: 10.1038/s41598-020-59292-w. Sci Rep. 2020. PMID: 32080241 Free PMC article.
References
- Pollitzer R. 1959. Cholera. World Health Organization, Geneva, Switzerland
- Pan American Health Organization Monday, July 25, 2011,. Cholera and post-earth-quake response in Haiti. Health Cluster Bulletin 26 http://new.paho.org/blogs/haiti/?p=2043
- Centers for Disease Control and Prevention 2010. Update on cholera—Haiti, Dominican Republic, and Florida. MMWR Morb. Mortal. Wkly. Rep. 59:1637–1641 - PubMed
- Butler D. 2010. Cholera tightens grip on Haiti. Nature 468:2 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical