Structural and mechanistic commonalities of amyloid-β and the prion protein - PubMed (original) (raw)
Review
Structural and mechanistic commonalities of amyloid-β and the prion protein
Bianca Da Costa Dias et al. Prion. 2011 Jul-Sep.
Abstract
Amyloid β (Aβ) is a major causative agent of Alzheime disease. This neurotoxic peptide is generated as a result of the cleavage of the Amyloid-Precursor-Protein (APP) by the action of beta secretase and gamma secretase. The neurotoxicity was previously thought to be the result of aggregation. However, recent studies suggest that the interaction of Aβ with numerous cell surface receptors such as N-methyl-D-aspartate (NMDA), receptor for advanced glycosylation end products (RAGE), P75 neurotrophin receptor (P75NTR) as well as cell surface proteins such as the cellular prion protein (PrP(c) ) and heparan sulfate proteoglycans (HSPG) strongly enhances Aβ induced apoptosis and thereby contributes to neurotoxicity. This review focuses on the molecular mechanism resulting in Aβ-shedding as well as Aβ-induced apoptotic processes, genetic risk factors for familial Alzheimer disease and interactions of Aβ with cell surface receptors and proteins, with particular emphasis on the cellular prion protein. Furthermore, comparisons are drawn between Alzheimer disease and prion disorders and the role of laminin, an extracellular matrix protein, glycosaminoglycans and the 37 kDa/67 kDa laminin receptor (LRP/LR) have been highlighted with regards to both neurodegenerative diseases.
Figures
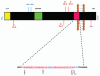
Figure 1
Schematic representation of the APP and the enzymatic cleavage sites located within the amyloid β sequence. (A) This transmembrane protein may be present in multiple isoforms APP695, APP751 and APP770, the latter is represented here. The regions of interest depicted in the diagram are: A signal peptide (yellow box) comprising 17 amino acid residues which ensures the protein is correctly transported to the cell surface; a 56 amino acid Kunitz-type serine protease inhibitor domain (KPI-green box) and the Aβ sequence. In addition, the sites of post-translational modifications such as N- and O-linked sugars (NCH2O and OCH2O), phosphate (PO4) and sulphate (SO4) groups are shown. (B) The 40–42 amino acid Aβ sequence is highlighted above-the first 28 amino acid residues are polar and located on the extracellular domain of APP whilst the remaining residues are located within the 23 aa APP transmembrane domain and are non-polar. The enzymatic cleavage sites of β secretase, α secretase and γ secretase are depicted (Adapted from reference 112).
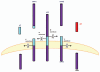
Figure 2
The proteolytic processing of the APP and its cleavage products. The amyloid precursor protein may be metabolized through two pathways. The first, depicted on the left, is termed the non-amyloidogenic pathways. This pathway involves the enzymatic cleavage by an α-secretase (presumably a member of the ADAM family) after residue 687, followed by γ-secretase mediated cleavage of the remaining carboxyl terminal fragment (CTF83), generating sAPPα, p3 and AICD (not shown), respectively. The amyloidogenic pathway (right) entails β-secretase mediated cleavage after residue 671, thereby releasing sAPPβ and the resultant CTF99 is cleaved by γ-secretase which results in Aβ shedding. The miscellaneous receptor, which may influence the process or serve as a receptor for either Aβ cleavage products, is hypothesized to be the 37 kDa/67 kDa Laminin Receptor Precursor/Laminin Receptor (LRP/LR) (adapted from reference 32).
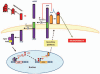
Figure 3
Feedback loop for the prion protein (PrPc) mediated regulation of APP. The amyloid intracellular domain (AICD) amyloidogenic APP cleavage product, indirectly upregulates prion protein (PrPc) expression through p53 gene activation. PrPc consequently hampers β-secretase (BACE1) activity thereby reducing APP amyloidogenic processing and Aβ synthesis. In the presence of Aβ-oligomers, which preferentially bind to PrPc, PrPc is unable to inhibit β-secretase (BACE1) activity. This reduces the degree of regulatory control exerted on the amyloidogenic process resulting in increased levels of potentially toxic Aβ oligomers (adapted from reference 81).
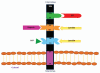
Figure 4
Schematic representation of the the functional domains of the 37 kDa/67 kDa Laminin Receptor Precursor/Laminin Receptor. This receptor, which is 295 amino acids in length, may be located at the cell surface, in the cytoplasm and the nucleus and displays different functional roles in each. A cell-surface associated form of the multi-functional protein is depicted here. The transmembrane domain of the receptor is located between amino acid residues LRP86–101. In this location the protein functions primarily as a receptor and encompasses four defined ligand-binding domains, including a prion protein binding domain (LRP161–180) and two laminin binding domains (LRP160–180 and LRP205–229), the latter functions as a heparin binding domain as well and an IgG-antibody binding domain (LRP272–280) (adapted from reference 32).
Figure 5
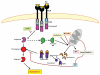
A schematic representation of the two classical apoptotic pathways in mammalian cells. The extrinsic is triggered at the cell surface by ligand (CD95L, FASL, TNFα) binding to death receptors (CD95, FASR, TNFR) and the ultimate formation of a death inducing signaling complex (DISC). Conversely the intrinsic pathway involves alterations in mitochondrion permeability as a result of intracellular signals such as DNA damage and oxidative stress. The activation of the aforementioned pathways leads to the cleavage and activation of initiator caspases 8 and caspases 9 respectively. These in turn activate the effector caspases 3 which facilitates DNA fragmentation and cytoskeletal protein degradation-leading to the morphological and physical features characteristic of apoptotic cells (adapted from reference 96).
Similar articles
- Binding sites of amyloid beta-peptide in cell plasma membrane and implications for Alzheimer's disease.
Verdier Y, Penke B. Verdier Y, et al. Curr Protein Pept Sci. 2004 Feb;5(1):19-31. doi: 10.2174/1389203043486937. Curr Protein Pept Sci. 2004. PMID: 14965318 Review. - Anti-LRP/LR specific antibodies and shRNAs impede amyloid beta shedding in Alzheimer's disease.
Jovanovic K, Gonsalves D, Da Costa Dias B, Moodley K, Reusch U, Knackmuss S, Penny C, Weinberg MS, Little M, Weiss SF. Jovanovic K, et al. Sci Rep. 2013;3:2699. doi: 10.1038/srep02699. Sci Rep. 2013. PMID: 24048412 Free PMC article. - Amyloid beta-peptide interactions with neuronal and glial cell plasma membrane: binding sites and implications for Alzheimer's disease.
Verdier Y, Zarándi M, Penke B. Verdier Y, et al. J Pept Sci. 2004 May;10(5):229-48. doi: 10.1002/psc.573. J Pept Sci. 2004. PMID: 15160835 Review. - Site-specific blockade of RAGE-Vd prevents amyloid-beta oligomer neurotoxicity.
Sturchler E, Galichet A, Weibel M, Leclerc E, Heizmann CW. Sturchler E, et al. J Neurosci. 2008 May 14;28(20):5149-58. doi: 10.1523/JNEUROSCI.4878-07.2008. J Neurosci. 2008. PMID: 18480271 Free PMC article. - Ectodomain shedding of the receptor for advanced glycation end products: a novel therapeutic target for Alzheimer's disease.
Zhang L, Postina R, Wang Y. Zhang L, et al. Cell Mol Life Sci. 2009 Dec;66(24):3923-35. doi: 10.1007/s00018-009-0121-4. Cell Mol Life Sci. 2009. PMID: 19672558 Free PMC article. Review.
Cited by
- Looking into laminin receptor: critical discussion regarding the non-integrin 37/67-kDa laminin receptor/RPSA protein.
DiGiacomo V, Meruelo D. DiGiacomo V, et al. Biol Rev Camb Philos Soc. 2016 May;91(2):288-310. doi: 10.1111/brv.12170. Epub 2015 Jan 28. Biol Rev Camb Philos Soc. 2016. PMID: 25630983 Free PMC article. Review. - Amyloid-beta Alzheimer targets - protein processing, lipid rafts, and amyloid-beta pores.
Arbor SC, LaFontaine M, Cumbay M. Arbor SC, et al. Yale J Biol Med. 2016 Mar 24;89(1):5-21. eCollection 2016 Mar. Yale J Biol Med. 2016. PMID: 27505013 Free PMC article. Review. - β-Cleavage of the prion protein in the human eye: Implications for the spread of infectious prions and human ocular disorders.
Chaudhary S, Ashok A, Wise AS, Rana NA, Kritikos AE, Lindner E, Singh N. Chaudhary S, et al. Exp Eye Res. 2021 Nov;212:108787. doi: 10.1016/j.exer.2021.108787. Epub 2021 Oct 7. Exp Eye Res. 2021. PMID: 34624335 Free PMC article. - Modulation of γ-secretase activity by multiple enzyme-substrate interactions: implications in pathogenesis of Alzheimer's disease.
Svedružić ZM, Popović K, Smoljan I, Sendula-Jengić V. Svedružić ZM, et al. PLoS One. 2012;7(3):e32293. doi: 10.1371/journal.pone.0032293. Epub 2012 Mar 30. PLoS One. 2012. PMID: 22479317 Free PMC article. - Receptor for Advanced Glycation End Products (RAGE): A Pivotal Hub in Immune Diseases.
Yue Q, Song Y, Liu Z, Zhang L, Yang L, Li J. Yue Q, et al. Molecules. 2022 Aug 2;27(15):4922. doi: 10.3390/molecules27154922. Molecules. 2022. PMID: 35956875 Free PMC article. Review.
References
- Mount C, Downton C. Alzheimer disease: progress or profit? Nat Med. 2006;12:780–784. - PubMed
- Palmer AM. Neuroprotective therapeutics for Alzheimer's disease: progress and prospects. Trends Pharmacol Sci. 2011;32:141–147. - PubMed
- Selkoe DJ. Alzheimer's disease: genes, proteins and therapy. Physiol Rev. 2001;81:741–766. - PubMed
- Burns A, Iliffe S. Alzheimer's disease. BMJ. 2009;338:158. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous