The RND-family transporter, HpnN, is required for hopanoid localization to the outer membrane of Rhodopseudomonas palustris TIE-1 - PubMed (original) (raw)
The RND-family transporter, HpnN, is required for hopanoid localization to the outer membrane of Rhodopseudomonas palustris TIE-1
David M Doughty et al. Proc Natl Acad Sci U S A. 2011.
Abstract
Rhodopseudomonas palustris TIE-1 is a gram-negative bacterium that produces structurally diverse hopanoid lipids that are similar to eukaryotic steroids. Its genome encodes several homologues to proteins involved in eukaryotic steroid trafficking. In this study, we explored the possibility that two of these proteins are involved in intracellular hopanoid transport. R. palustris has a sophisticated membrane system comprising outer, cytoplasmic, and inner cytoplasmic membranes. It also divides asymmetrically, producing a mother and swarmer cell. We deleted genes encoding two putative hopanoid transporters that belong to the resistance-nodulation-cell division superfamily. Phenotypic analyses revealed that one of these putative transporters (HpnN) is essential for the movement of hopanoids from the cytoplasmic to the outer membrane, whereas the other (Rpal_4267) plays a minor role. C(30) hopanoids, such as diploptene, are evenly distributed between mother and swarmer cells, whereas hpnN is required for the C(35) hopanoid, bacteriohopanetetrol, to remain localized to the mother cell type. Mutant cells lacking HpnN grow like the WT at 30 °C but slower at 38 °C. Following cell division at 38 °C, the ΔhpnN cells remain connected by their cell wall, forming long filaments. This phenotype may be attributed to hopanoid mislocalization because a double mutant deficient in both hopanoid biosynthesis and transport does not form filaments. However, the lack of hopanoids severely compromises cell growth at higher temperatures more generally. Because hopanoid mutants only manifest a strong phenotype under certain conditions, R. palustris is an attractive model organism in which to study their transport and function.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
Structures of cholesterol and hopanoid lipids and arrangement of hopanoid biosynthetic genes on the chromosome of R. palustris TIE-1. (A) Comparison of cholesterol and hopanoids. The “unextended” hopanoids (C30 or C31) are generally considered to be intermediates in the biosynthesis of “extended” (C35+) hopanoids. When R1 is H, the unextended hopanoid is called diplopterol. When R1 is H and R2 is OH, the extended hopanoid is called BHT (12). (B) Graphical depiction of the hopanoid biosynthetic gene cluster of R. palustris TIE-1 labeled by gene number and names. Genes known to be essential to hopanoid biosynthesis are shown in black, and genes encoding putative hopanoid transporters are shown in gray.
Fig. 2.
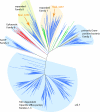
Unrooted phylogeny of the RND superfamily showing the eight recognized families of RND transporters (
) (22). The phylogenetic associations of Rpal_4254 and Rpal_4267 within these families are indicated by the arrows. (Scale bar: amino acid substitutions per site.)
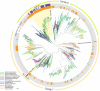
Fig. 3.
Maximum likelihood phylogeny of two families of RND transporters associated with hopanoid transport. Concentric rings depict co-occurrence of squalene hopene cyclase in the genome (inner ring) and proximity to genes known to be involved in hopanoid biosynthesis (outer ring). Nodes with bootstrap support greater than 95% are marked with a gray circle. For clarity, sequence names have been omitted and closely related clades have been collapsed; the full version is shown in
Fig. S1
.
Fig. 4.
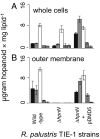
Whole cell (A) and outer membrane (B) hopanoid content of WT, ΔhpnN, and ΔhpnN strain complemented with pDMD5. Hopanoids are diploptene (black), 2-methyldiploptene (gray), BHT (white), 2-methylBHT (horizontal stripes), and bacteriohopaneaminotriol (vertical stripes). The error bars represent the SD from the mean of three independent cultures.
Fig. 5.
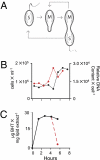
Physiology and hopanoid production in synchronized cultures of R. palustris. (A) Physical depiction of the cell cycle of R. palustris TIE-1. (B) Direct cell count (black line) and DNA content (red line) of cells in a representative synchronized culture (four independent cultures were analyzed). (C) BHT content of a representative synchronized culture (three independent cultures were analyzed). The black line follows BHT concentration in a swarmer cell as it grows into a mother cell. The red line indicates the BHT concentration of the new swarmer cell that is formed following one cell cycle.
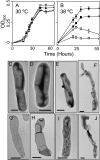
Fig. 6.
The effects of elevated temperature on cell growth and physiology. Growth curves conducted at 30 °C (A) and 38 °C (B) on the WT (●), ΔhpnN (■), Δshc (○), and ΔhpnNΔshc (□). Error bars represent the SD from the means of three independent cultures. Visualization of whole cell morphology in WT grown at 30 °C (C) and 38 °C (D) and of ΔhpnN grown at 30 °C (E) and 38 °C (F). Visualization of peptidoglycan sacculi in WT grown at 30 °C (G) and 38 °C (H) and of ΔhpnN grown at 30 °C (I) and 38 °C (I). (Scale bars: C_–_J, 0.5 μm.)
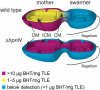
Fig. P1.
Partitioning of the extended hopanoid BHT within R. palustris TIE-1 as a function of the presence of the RND-transporter HpnN. BHT is primarily an outer membrane (OM) lipid in WT R. palustris, with only a small presence in the cytoplasmic membrane (CM) and inner cytoplasmic membrane (ICM). Interestingly, whereas BHT is present in mother cells of the WT, it is absent in swarmer cells. Deletion of the HpnN transporter traps BHT within the inner membranes, thus permitting hopanoids to enter swarmer cells. Cyan indicates membranes with no measurable BHT, yellow indicates the presence of small amounts of BHT [∼2–5 μg/mg total lipid extract (TLE)], and magenta indicates high concentrations of BHT (>10 μg /mg TLE).
Similar articles
- Transcriptome analysis of hopanoid deficient mutant of Rhodopseuodomonas palustris TIE-1.
Lodha TD, B I, Ch S, Ch V R. Lodha TD, et al. Microbiol Res. 2019 Jan;218:108-117. doi: 10.1016/j.micres.2018.10.009. Epub 2018 Oct 29. Microbiol Res. 2019. PMID: 30454652 - Identification and characterization of Rhodopseudomonas palustris TIE-1 hopanoid biosynthesis mutants.
Welander PV, Doughty DM, Wu CH, Mehay S, Summons RE, Newman DK. Welander PV, et al. Geobiology. 2012 Mar;10(2):163-77. doi: 10.1111/j.1472-4669.2011.00314.x. Epub 2012 Jan 4. Geobiology. 2012. PMID: 22221333 Free PMC article. - Crystal structures of the Burkholderia multivorans hopanoid transporter HpnN.
Kumar N, Su CC, Chou TH, Radhakrishnan A, Delmar JA, Rajashankar KR, Yu EW. Kumar N, et al. Proc Natl Acad Sci U S A. 2017 Jun 20;114(25):6557-6562. doi: 10.1073/pnas.1619660114. Epub 2017 Jun 5. Proc Natl Acad Sci U S A. 2017. PMID: 28584102 Free PMC article. - Hopanoid lipids: from membranes to plant-bacteria interactions.
Belin BJ, Busset N, Giraud E, Molinaro A, Silipo A, Newman DK. Belin BJ, et al. Nat Rev Microbiol. 2018 May;16(5):304-315. doi: 10.1038/nrmicro.2017.173. Epub 2018 Feb 19. Nat Rev Microbiol. 2018. PMID: 29456243 Free PMC article. Review. - RND efflux pumps in Gram-negative bacteria; regulation, structure and role in antibiotic resistance.
Colclough AL, Alav I, Whittle EE, Pugh HL, Darby EM, Legood SW, McNeil HE, Blair JM. Colclough AL, et al. Future Microbiol. 2020 Jan;15:143-157. doi: 10.2217/fmb-2019-0235. Epub 2020 Feb 19. Future Microbiol. 2020. PMID: 32073314 Review.
Cited by
- Membrane lipids in Agrobacterium tumefaciens: biosynthetic pathways and importance for pathogenesis.
Aktas M, Danne L, Möller P, Narberhaus F. Aktas M, et al. Front Plant Sci. 2014 Mar 26;5:109. doi: 10.3389/fpls.2014.00109. eCollection 2014. Front Plant Sci. 2014. PMID: 24723930 Free PMC article. Review. - Lack of Methylated Hopanoids Renders the Cyanobacterium Nostoc punctiforme Sensitive to Osmotic and pH Stress.
Garby TJ, Matys ED, Ongley SE, Salih A, Larkum AWD, Walter MR, Summons RE, Neilan BA. Garby TJ, et al. Appl Environ Microbiol. 2017 Jun 16;83(13):e00777-17. doi: 10.1128/AEM.00777-17. Print 2017 Jul 1. Appl Environ Microbiol. 2017. PMID: 28455341 Free PMC article. - Patched1 and Patched2 inhibit Smoothened non-cell autonomously.
Roberts B, Casillas C, Alfaro AC, Jägers C, Roelink H. Roberts B, et al. Elife. 2016 Aug 23;5:e17634. doi: 10.7554/eLife.17634. Elife. 2016. PMID: 27552050 Free PMC article. - Novel sterol binding domains in bacteria.
Zhai L, Bonds AC, Smith CA, Oo H, Chou JC, Welander PV, Dassama LMK. Zhai L, et al. Elife. 2024 Feb 8;12:RP90696. doi: 10.7554/eLife.90696. Elife. 2024. PMID: 38329015 Free PMC article.
References
- Brocks JJ, Buick R, Logan GA, Summons RE. Composition and syngeneity of molecular fossils from the 2.78 to 2.45 billion-year-old Mount Bruce Subgroup, Pibara Craton, Western Australia. Geochim Cosmochim Acta. 2003;67:4289–4319.
- Eigenbrode JL. Fossil lipids for life-detection: A case study from the early earth record. Space Sci Rev. 2008;135:161–185.
- Waldbauer JR, Sherman LS, Sumner DY, Summons RE. Late Archean molecular fossils from the Transvaal Supergroup record the antiquity of microbial diversity and aerobiosis. Precambrian Res. 2009;169:28–47.
- Eaton S. Multiple roles for lipids in the Hedgehog signalling pathway. Nat Rev Mol Cell Biol. 2008;9:437–445. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous